
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,172,807 – 4,172,964 |
| Length | 157 |
| Max. P | 0.954054 |

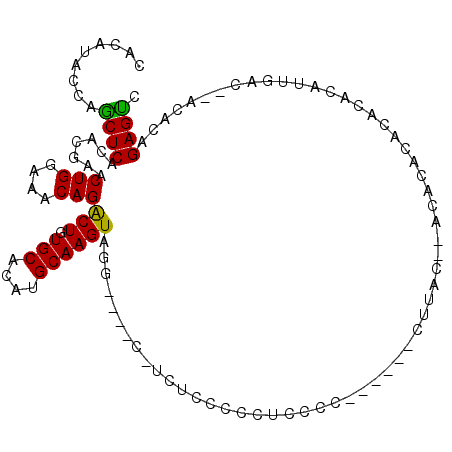
| Location | 4,172,807 – 4,172,906 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.83 |
| Mean single sequence MFE | -19.43 |
| Consensus MFE | -15.08 |
| Energy contribution | -14.44 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4172807 99 - 23771897 CACAUACCAACUCACACGAACUGGAAACAGACUGUGCACAUGCAAGUAGG----C-UCUCCCCCUCCCA------CUUACCCACACACACACACAUUGAC--ACACAGAGUC .........((((.((((..(((....)))..)))).......((((.((----.-.........)).)------)))......................--.....)))). ( -14.40) >DroSec_CAF1 18471 102 - 1 CACAUACCAGCUCACACGAACUGGAAACAGACUGUGCACAUGCAAGUGGAGAGCCUACUCUCCCUCCCC------CUUAC--ACACACACACACAUUGGC--ACACAGAGUC .........((((.......(((....)))..(((((.......((.((((((....))))))))....------.....--.......((.....))))--)))..)))). ( -24.60) >DroSim_CAF1 22995 97 - 1 CACAUACCAGCUCACACGAACUGGAAACAGACUGUGCACAUGCAAGUAGG----C-UCUCCCCCUCCCC------CUUAC--ACACACACACACAUUGAC--ACAGAGAGUC ....(((..((...((((..(((....)))..)))).....))..)))((----(-((((.........------.....--..................--...))))))) ( -17.75) >DroEre_CAF1 18914 105 - 1 CACAUACCAGCUCACACGAACUGGAAACAGGCUGUGCACAUGCAAGUAGG----C-UCUCCCCUCCCCCUUCCUUCUUAC--ACACACACACACACUGACAUACACAGAGCC .........((((.((((..(((....)))..))))....((.(((.(((----.-...............))).))).)--)........................)))). ( -18.89) >DroYak_CAF1 22877 103 - 1 CACAUACCAGCUCACACGAACUGGAAACAGGCUGUGCACAUGCAAGUAGG----C-UCUCCCCCUACCCCUCC--CCAAC--CCCCUUACACACACUGACAUACACAGAGCC .........((((.((((..(((....)))..)))).........(((((----.-......)))))......--.....--.........................)))). ( -21.50) >consensus CACAUACCAGCUCACACGAACUGGAAACAGACUGUGCACAUGCAAGUAGG____C_UCUCCCCCUCCCC______CUUAC__ACACACACACACAUUGAC__ACACAGAGUC .........((((.......(((....)))(((.(((....))))))............................................................)))). (-15.08 = -14.44 + -0.64)

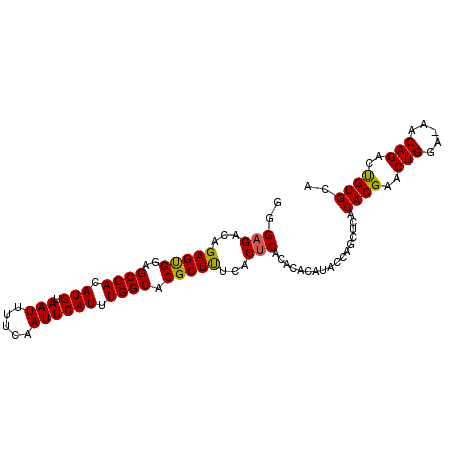
| Location | 4,172,868 – 4,172,964 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -22.10 |
| Energy contribution | -21.85 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4172868 96 - 23771897 GGGAGACAGAGUGGAGCCACAUGUAAUUUUCAAUUCAUUUGGCACGCUUUCACUCACACACAUACCAACUCACACGAACUGGA-AACAGACUGUGCA ..(((..((((((..((((.(((.(((.....)))))).)))).))))))..))).................((((..(((..-..)))..)))).. ( -24.80) >DroSec_CAF1 18535 96 - 1 GGGAGACAGAGUGGAGCCACAUGUAAUUUUCAAUUCAUUUGGCACGCUUUCACUCACACACAUACCAGCUCACACGAACUGGA-AACAGACUGUGCA ..(((..((((((..((((.(((.(((.....)))))).)))).))))))..))).................((((..(((..-..)))..)))).. ( -24.80) >DroSim_CAF1 23054 96 - 1 GGGAGACAGAGUGGAGCCACAUGUAAUUUUCAAUUCAUUUGGCACGCUUUCACUCACACACAUACCAGCUCACACGAACUGGA-AACAGACUGUGCA ..(((..((((((..((((.(((.(((.....)))))).)))).))))))..))).................((((..(((..-..)))..)))).. ( -24.80) >DroEre_CAF1 18981 96 - 1 GGGAGACAGAGUGGAGCCACAUGUAAUUUUCAAUUCAUUUGGCACGCUUUCACUCACACACAUACCAGCUCACACGAACUGGA-AACAGGCUGUGCA ..(((..((((((..((((.(((.(((.....)))))).)))).))))))..))).................((((..(((..-..)))..)))).. ( -26.80) >DroYak_CAF1 22942 96 - 1 GUGAGACAGAGUGGAGCCACAUGUAAUUUUCAAUUCAUUUGGCACGCUUUCACUCACACACAUACCAGCUCACACGAACUGGA-AACAGGCUGUGCA (((((..((((((..((((.(((.(((.....)))))).)))).))))))..)))))...............((((..(((..-..)))..)))).. ( -31.10) >DroAna_CAF1 19222 93 - 1 GGGAGACAGAGCGGAGCCACAUGUAAUUUUCAAUUCAUUUGGCACGCUCGGACACACACACACAC--GCUCACACG--CUGGAAAACAGGCCGUGCA ..(((.(.(((((..((((.(((.(((.....)))))).)))).)))))(......)........--)))).((((--(((.....)))..)))).. ( -25.00) >consensus GGGAGACAGAGUGGAGCCACAUGUAAUUUUCAAUUCAUUUGGCACGCUUUCACUCACACACAUACCAGCUCACACGAACUGGA_AACAGACUGUGCA ..(((...(((((..((((.(((.(((.....)))))).)))).)))))...))).................((((..(((.....)))..)))).. (-22.10 = -21.85 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:47 2006