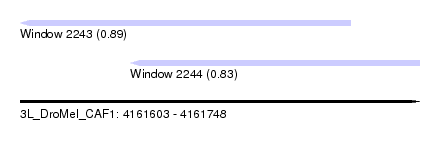
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,161,603 – 4,161,748 |
| Length | 145 |
| Max. P | 0.886751 |

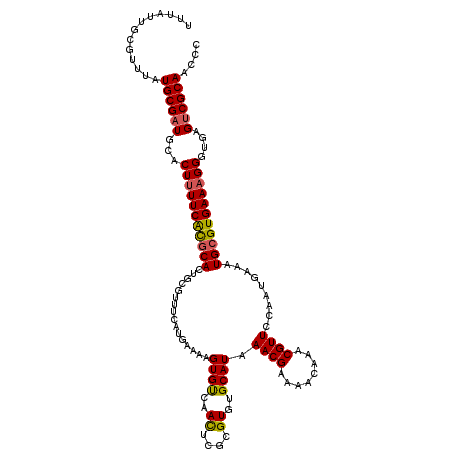
| Location | 4,161,603 – 4,161,723 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -26.56 |
| Energy contribution | -26.44 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4161603 120 - 23771897 UUUAUUGCGUUUAUGCGAUGCACUUUUCACGCACUGCGUUUCAUGAAAAGUGUCAACACGCGUGUGCAUAAACGAAAACAAACGUUCCAAUGAAAUGCGUGAAAGGGUGAGUCGCAACCC ....((((((((((.(...(((((((((((((...)))).....)))))))))...(((((((...(((.((((........))))...)))..)))))))...).))))).)))))... ( -35.80) >DroSec_CAF1 7359 120 - 1 UUUAUUGUGUUUAUGCGAUGAACUUUUCACACAUUCCGUUUCAUGAAAAGUGUCAACUCGCGUGUGCAUAAACGAAAACAAACGUUCCAAUGAAAUGCGUGAAAGGGUGAGUCGCAACCC .............((((((...((((((((.((((.((((.((((...(((....)))..))))......((((........))))..)))).)))).))))))))....)))))).... ( -29.40) >DroSim_CAF1 11688 120 - 1 UUUAUUGUGUUUAUGCGAUGCACUUUUCACGCACUGCGUUUCAUGAAAAGUGUCAACUCGCGUGUGCAUAAACGAAAACAAACGUUCCAAUAAAAUGCGUGAAAGGGUGAGUCGCAACCC .............((((((.(((((((((((((.((((...((((...(((....)))..))))))))..((((........)))).........))))))))).)))).)))))).... ( -33.90) >DroEre_CAF1 7443 106 - 1 UUUACUGCGUUUAUGCGUUGCACUUUUCGCGCA--------------GAGUGCCAAUUCGUGUGUGCAUAAACGAAAACAAACGUUUAAAUGAAAUGCGUGAAAGGGCGAGUCGCAACCC .............((((((((.(((((((((((--------------..(..(((.....)).)..).((((((........)))))).......)))))))))))))))..)))).... ( -32.40) >DroYak_CAF1 11343 120 - 1 UUUACUGCGUUUAUGCGAUGCACUUUUCAUGCAUUUACUUUCCUGAGAAGUGCCAACUCGCGUGUGCAUAAACGAAAGCAAACGUUCGAGUAAAAUGCGUGAACGGGUGAGUCGCAACCC .............((((((.(((((((((((((((((((((.....)))))....((((((((.(((..........))).)))..))))).))))))))))).))))).)))))).... ( -39.10) >consensus UUUAUUGCGUUUAUGCGAUGCACUUUUCACGCACUGCGUUUCAUGAAAAGUGUCAACUCGCGUGUGCAUAAACGAAAACAAACGUUCCAAUGAAAUGCGUGAAAGGGUGAGUCGCAACCC .............((((((...(((((((((((................((((..((....))..)))).((((........)))).........)))))))))))....)))))).... (-26.56 = -26.44 + -0.12)

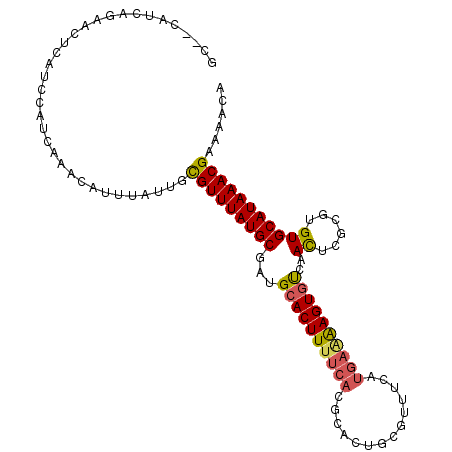
| Location | 4,161,643 – 4,161,748 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -18.60 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4161643 105 - 23771897 GC--CAUCAGAACGCAUCCAUCAAACAUUUAUUGCGUUUAUGCGAUGCACUUUUCACGCACUGCGUUUCAUGAAAAGUGUCAACACGCGUGUGCAUAAACGAAAACA ((--(((..(((((((................)))))))..(((.((((((((((((((...)))).....)))))))).))...)))))).))............. ( -25.39) >DroSec_CAF1 7399 105 - 1 GC--CAUCAGAACUCAUCCAUCAAACAUUUAUUGUGUUUAUGCGAUGAACUUUUCACACAUUCCGUUUCAUGAAAAGUGUCAACUCGCGUGUGCAUAAACGAAAACA ..--...........................((((((.((((((((((((((((((.((.....))....)))))))).)))..))))))).))))))......... ( -21.50) >DroSim_CAF1 11728 105 - 1 GC--CAUCAGAACUCAUCCAUCAAACAUUUAUUGUGUUUAUGCGAUGCACUUUUCACGCACUGCGUUUCAUGAAAAGUGUCAACUCGCGUGUGCAUAAACGAAAACA ..--...........................((((((.(((((((((((((((((((((...)))).....)))))))).))..))))))).))))))......... ( -26.00) >DroEre_CAF1 7483 91 - 1 GC--AAUCAGAACUCAUCCAUCAAACAUUUACUGCGUUUAUGCGUUGCACUUUUCGCGCA--------------GAGUGCCAAUUCGUGUGUGCAUAAACGAAAACA ..--..............................(((((((((...((((..((.((((.--------------..)))).))...))))..)))))))))...... ( -18.50) >DroYak_CAF1 11383 107 - 1 ACUCUAUUAAAACUCAUCCAUCAAACAUUUACUGCGUUUAUGCGAUGCACUUUUCAUGCAUUUACUUUCCUGAGAAGUGCCAACUCGCGUGUGCAUAAACGAAAGCA ..................................(((((((((...((((((((((.(.(......).).))))))))))..((......)))))))))))...... ( -23.40) >consensus GC__CAUCAGAACUCAUCCAUCAAACAUUUAUUGCGUUUAUGCGAUGCACUUUUCACGCACUGCGUUUCAUGAAAAGUGUCAACUCGCGUGUGCAUAAACGAAAACA ..................................(((((((((...((((((((((..............))))))))))..((......)))))))))))...... (-18.60 = -18.68 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:42 2006