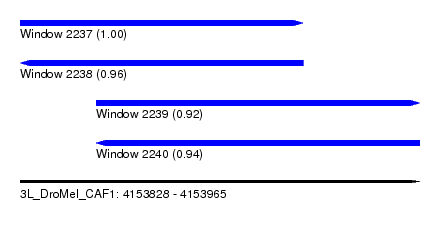
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,153,828 – 4,153,965 |
| Length | 137 |
| Max. P | 0.999424 |

| Location | 4,153,828 – 4,153,925 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 70.97 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.46 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4153828 97 + 23771897 -UAAUG-------GGAAAUAU---UGGCCAUUUGAAAAGCACCUGCUGUGCUGAGUGACCGCACUUCG---------CUCGUCAAGUGUGGUAUUUUUUAGUACGUUUCGGCGCUUA -.((((-------(.......---...)))))....((((.((.((.((((((((..(((((((((..---------......)))))))))....))))))))))...)).)))). ( -30.40) >DroSim_CAF1 3415 108 + 1 UAAAUUUAAGCAUGUAAGCACUUGUUACUUUUUGAAAAGCACCCGCUGUGCUGAGUGACCGCACUUUG---------CUCGGCAAGUGUGGUAUUUUUUAGUACUUUUCGGCGCUUA ....((((((...((((.(....)))))..))))))((((.((....((((((((..(((((((((.(---------(...)))))))))))....)))))))).....)).)))). ( -31.20) >DroYak_CAF1 3585 114 + 1 ---AUUGAAAUAUUUACACACUUGUUCCUAUUUGAAAAGCACCUGCUGUACUGAGUGACCGCAUUCCGUAAUGCUACCUCGCAAGACGUGGUAUUUUUUAGUACAUUUCGGCGCUCG ---.((.(((((...(((....)))...))))).)).(((.((...(((((((((((....)))))...((((((((.((....)).))))))))....))))))....)).))).. ( -28.20) >consensus __AAUU_AA__AUGUAAACACUUGUUACUAUUUGAAAAGCACCUGCUGUGCUGAGUGACCGCACUUCG_________CUCGCCAAGUGUGGUAUUUUUUAGUACAUUUCGGCGCUUA ....................................((((.((...(((((((((..(((((((((.................)))))))))....)))))))))....)).)))). (-23.12 = -23.46 + 0.34)

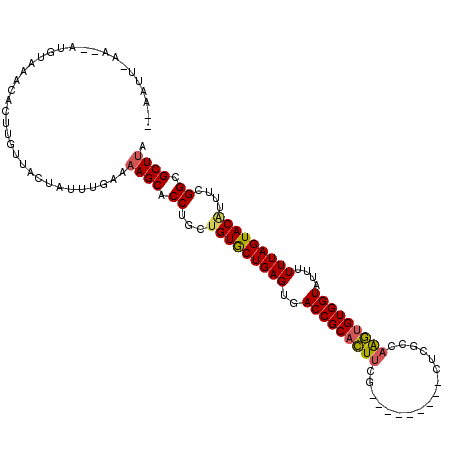
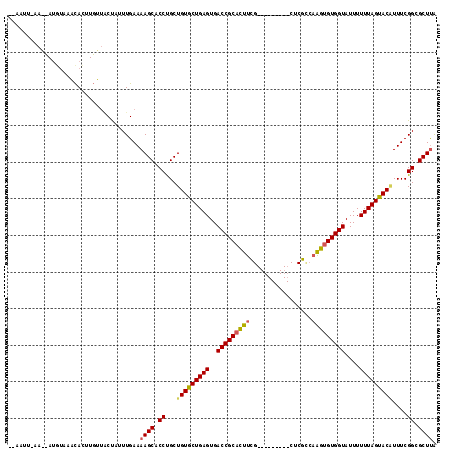
| Location | 4,153,828 – 4,153,925 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.97 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -16.25 |
| Energy contribution | -15.26 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4153828 97 - 23771897 UAAGCGCCGAAACGUACUAAAAAAUACCACACUUGACGAG---------CGAAGUGCGGUCACUCAGCACAGCAGGUGCUUUUCAAAUGGCCA---AUAUUUCC-------CAUUA- .(((((((.................(((.(((((......---------..))))).)))......((...)).)))))))....(((((...---.......)-------)))).- ( -20.50) >DroSim_CAF1 3415 108 - 1 UAAGCGCCGAAAAGUACUAAAAAAUACCACACUUGCCGAG---------CAAAGUGCGGUCACUCAGCACAGCGGGUGCUUUUCAAAAAGUAACAAGUGCUUACAUGCUUAAAUUUA ((((((..((((((((((.......(((.(((((((...)---------).))))).)))......((...)).))))))))))...(((((.....)))))...))))))...... ( -28.40) >DroYak_CAF1 3585 114 - 1 CGAGCGCCGAAAUGUACUAAAAAAUACCACGUCUUGCGAGGUAGCAUUACGGAAUGCGGUCACUCAGUACAGCAGGUGCUUUUCAAAUAGGAACAAGUGUGUAAAUAUUUCAAU--- .(((((((....((((((......((((.((.....)).))))(((((....)))))........))))))...))))))).............((((((....))))))....--- ( -29.90) >consensus UAAGCGCCGAAAAGUACUAAAAAAUACCACACUUGACGAG_________CGAAGUGCGGUCACUCAGCACAGCAGGUGCUUUUCAAAUAGCAACAAGUGUUUACAU__UU_AAUU__ .(((((((.................(((.(((((.................))))).)))......((...)).))))))).................................... (-16.25 = -15.26 + -0.99)

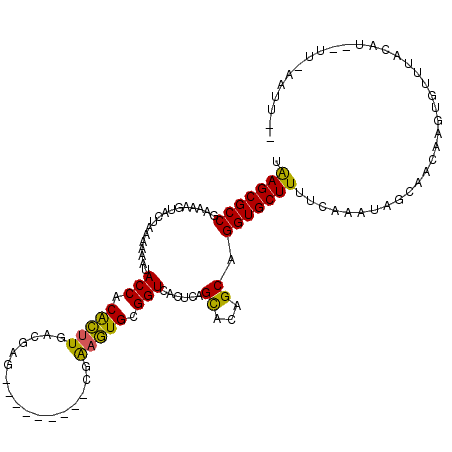

| Location | 4,153,854 – 4,153,965 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -42.53 |
| Consensus MFE | -32.95 |
| Energy contribution | -33.73 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
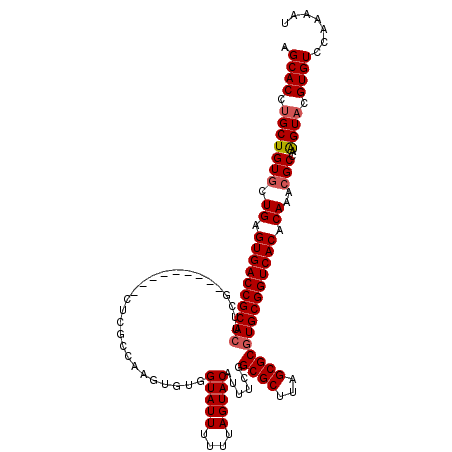
>3L_DroMel_CAF1 4153854 111 + 23771897 AGCACCUGCUGUGCUGAGUGACCGCACUUCG---------CUCGUCAAGUGUGGUAUUUUUUAGUACGUUUCGGCGCUUAGCGCGUGCGGUCACACAAACGCCAGGUACGUGUCCAAAAU .(.(((((.(((..((.((((((((((..((---------(((((((((((((.((.....)).))))))).))))...)))).)))))))))).)).))).))))).)........... ( -43.60) >DroSim_CAF1 3452 111 + 1 AGCACCCGCUGUGCUGAGUGACCGCACUUUG---------CUCGGCAAGUGUGGUAUUUUUUAGUACUUUUCGGCGCUUAGCGCGUGCGGUCACACAAACGCCAAGUACGUGUCCAAAAU .((((.((((((((((((..(((((((((.(---------(...)))))))))))..............))))))))..)))).))))((.(((....((.....))..))).))..... ( -39.99) >DroYak_CAF1 3619 120 + 1 AGCACCUGCUGUACUGAGUGACCGCAUUCCGUAAUGCUACCUCGCAAGACGUGGUAUUUUUUAGUACAUUUCGGCGCUCGGCGCCUGCGGUCACACAAACGCCAAGUACGUGUCCAAAAU .((((.((((....((.(((((((((......((((((((.((....)).))))))))..............(((((...)))))))))))))).)).......)))).))))....... ( -44.00) >consensus AGCACCUGCUGUGCUGAGUGACCGCACUUCG_________CUCGCCAAGUGUGGUAUUUUUUAGUACAUUUCGGCGCUUAGCGCGUGCGGUCACACAAACGCCAAGUACGUGUCCAAAAU .((((.(((((((.((.((((((((((..........................(((((....)))))......((((...)))))))))))))).))..)))..)))).))))....... (-32.95 = -33.73 + 0.78)


| Location | 4,153,854 – 4,153,965 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -41.83 |
| Consensus MFE | -35.57 |
| Energy contribution | -35.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
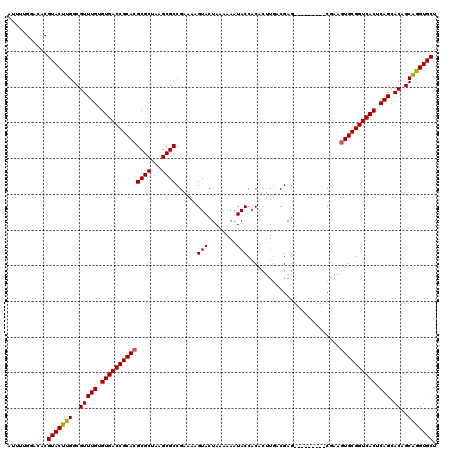
>3L_DroMel_CAF1 4153854 111 - 23771897 AUUUUGGACACGUACCUGGCGUUUGUGUGACCGCACGCGCUAAGCGCCGAAACGUACUAAAAAAUACCACACUUGACGAG---------CGAAGUGCGGUCACUCAGCACAGCAGGUGCU ...........(((((((..(((((.((((((((((((((...))))((...(((.(.................))))..---------))..)))))))))).))).))..))))))). ( -40.93) >DroSim_CAF1 3452 111 - 1 AUUUUGGACACGUACUUGGCGUUUGUGUGACCGCACGCGCUAAGCGCCGAAAAGUACUAAAAAAUACCACACUUGCCGAG---------CAAAGUGCGGUCACUCAGCACAGCGGGUGCU ...........(((((((..(((((.((((((((((((.....))((((..((((...............))))..)).)---------)...)))))))))).))).))..))))))). ( -39.16) >DroYak_CAF1 3619 120 - 1 AUUUUGGACACGUACUUGGCGUUUGUGUGACCGCAGGCGCCGAGCGCCGAAAUGUACUAAAAAAUACCACGUCUUGCGAGGUAGCAUUACGGAAUGCGGUCACUCAGUACAGCAGGUGCU ...........(((((((..(((((.((((((((((((((...))))).....................(((..(((......)))..)))...))))))))).))).))..))))))). ( -45.40) >consensus AUUUUGGACACGUACUUGGCGUUUGUGUGACCGCACGCGCUAAGCGCCGAAAAGUACUAAAAAAUACCACACUUGACGAG_________CGAAGUGCGGUCACUCAGCACAGCAGGUGCU ...........(((((((..(((((.((((((((((((((...))))......(((........)))..........................)))))))))).))).))..))))))). (-35.57 = -35.47 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:37 2006