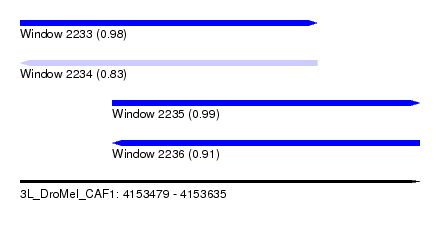
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,153,479 – 4,153,635 |
| Length | 156 |
| Max. P | 0.994603 |

| Location | 4,153,479 – 4,153,595 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -18.22 |
| Energy contribution | -17.78 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4153479 116 + 23771897 ---UCUCAUUCUGCUAAUUCAAAUCGAGCGGU-UUUAUUUCUGUUCUCCAUAACGCGAUCUGGCAACCUCUUCCCUUAAAAUGUUUCAUCCCUGGGAAGAGGGUUGCCUCUGCUUCAAAA ---......................((((((.-.......))))))........(((....((((((((((((((..................))))))).)))))))..)))....... ( -32.47) >DroSim_CAF1 3063 116 + 1 UGUUCUCAUUUUACCAAUUCAAAUCGAGCGGUCUUUACUAUUUUACUUUAUAACGCGAUCUGGCAACCUCUUCCCUU----UGUUUCAUCCCUGGAAUGAGGGUUGCCUCUGCUUCAAAU .........................((((((((.......................)))).((((((((.......(----..(((((....)))))..)))))))))...))))..... ( -20.51) >DroYak_CAF1 3249 120 + 1 UGUACUUUUCGUACAAAUUCAAAUCAAGCGGUCUUUACGCUUGUUCCUUAUACCGCGAUCUGGCAACCUCUUCUUUUAAAAUGCUUCAUCCCUGGAAAGAGGGUUGCCCUUUCUUCAAUU (((((.....))))).........((((((.......)))))).............((...((((((((..((((((...(((...))).....))))))))))))))...))....... ( -28.90) >consensus UGUUCUCAUUCUACAAAUUCAAAUCGAGCGGUCUUUACUAUUGUUCUUUAUAACGCGAUCUGGCAACCUCUUCCCUUAAAAUGUUUCAUCCCUGGAAAGAGGGUUGCCUCUGCUUCAAAU ...........................(((.......................))).....(((((((((((((...................)))).)).)))))))............ (-18.22 = -17.78 + -0.44)



| Location | 4,153,479 – 4,153,595 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -21.52 |
| Energy contribution | -23.30 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.828051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4153479 116 - 23771897 UUUUGAAGCAGAGGCAACCCUCUUCCCAGGGAUGAAACAUUUUAAGGGAAGAGGUUGCCAGAUCGCGUUAUGGAGAACAGAAAUAAA-ACCGCUCGAUUUGAAUUAGCAGAAUGAGA--- .......((...(((((((.(((((((.((((((...))))))..))))))))))))))((((((.((...((..............-.)))).))))))......)).........--- ( -33.56) >DroSim_CAF1 3063 116 - 1 AUUUGAAGCAGAGGCAACCCUCAUUCCAGGGAUGAAACA----AAGGGAAGAGGUUGCCAGAUCGCGUUAUAAAGUAAAAUAGUAAAGACCGCUCGAUUUGAAUUGGUAAAAUGAGAACA ..(..(..(((((((((((.((.((((..(.......).----...))))))))))))).((.((.(((..................))))).))..))))..)..)............. ( -22.87) >DroYak_CAF1 3249 120 - 1 AAUUGAAGAAAGGGCAACCCUCUUUCCAGGGAUGAAGCAUUUUAAAAGAAGAGGUUGCCAGAUCGCGGUAUAAGGAACAAGCGUAAAGACCGCUUGAUUUGAAUUUGUACGAAAAGUACA ..(..((((...(((((((.(((((...((((((...))))))....))))))))))))...)).............((((((.......)))))).))..)...(((((.....))))) ( -30.70) >consensus AUUUGAAGCAGAGGCAACCCUCUUUCCAGGGAUGAAACAUUUUAAGGGAAGAGGUUGCCAGAUCGCGUUAUAAAGAACAAAAGUAAAGACCGCUCGAUUUGAAUUAGUAAAAUGAGAACA ............(((((((.(((((((..(((((...)))))...))))))))))))))((((((((.......................)))..))))).................... (-21.52 = -23.30 + 1.78)

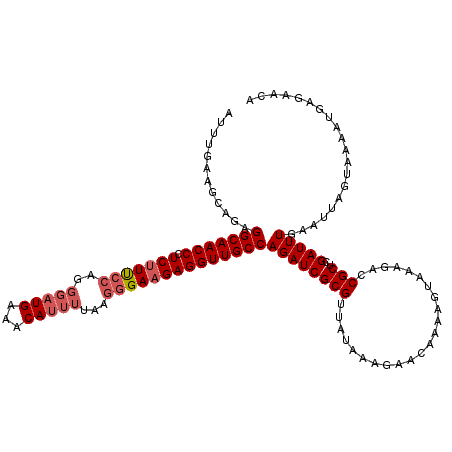

| Location | 4,153,515 – 4,153,635 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.61 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4153515 120 + 23771897 CUGUUCUCCAUAACGCGAUCUGGCAACCUCUUCCCUUAAAAUGUUUCAUCCCUGGGAAGAGGGUUGCCUCUGCUUCAAAAAAAUGGCGCGAAAUUUCAACAUAUCGUAAAUUCAAUAUAA ..............(((((..((((((((((((((..................))))))).)))))))..(((.(((......))).)))............)))))............. ( -30.67) >DroSim_CAF1 3103 112 + 1 UUUUACUUUAUAACGCGAUCUGGCAACCUCUUCCCUU----UGUUUCAUCCCUGGAAUGAGGGUUGCCUCUGCUUCAAAUAAGUGGCGCGAAAUUUCAACAUAUCGUCAG----AUAUAA .............((((....((((((((.......(----..(((((....)))))..))))))))).(..(((.....)))..)))))..........(((((....)----)))).. ( -27.71) >DroYak_CAF1 3289 107 + 1 UUGUUCCUUAUACCGCGAUCUGGCAACCUCUUCUUUUAAAAUGCUUCAUCCCUGGAAAGAGGGUUGCCCUUUCUUCAAUUAUUUGGCGCAAAAUUUGAACAUAUCAU------------- .(((((.......(((((...((((((((..((((((...(((...))).....))))))))))))))...))..(((....))))))........)))))......------------- ( -24.16) >consensus UUGUUCUUUAUAACGCGAUCUGGCAACCUCUUCCCUUAAAAUGUUUCAUCCCUGGAAAGAGGGUUGCCUCUGCUUCAAAUAAAUGGCGCGAAAUUUCAACAUAUCGU_A_____AUAUAA .............((((....(((((((((((((...................)))).)).)))))))......(((......))))))).............................. (-18.72 = -18.61 + -0.11)

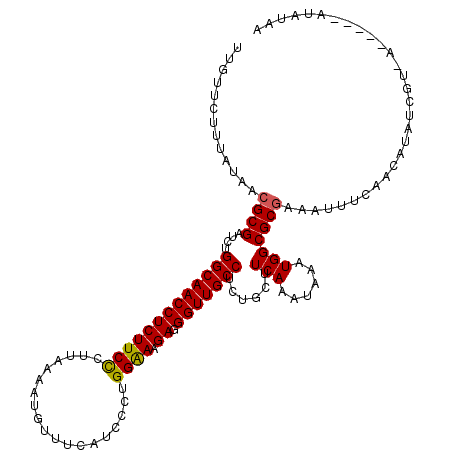
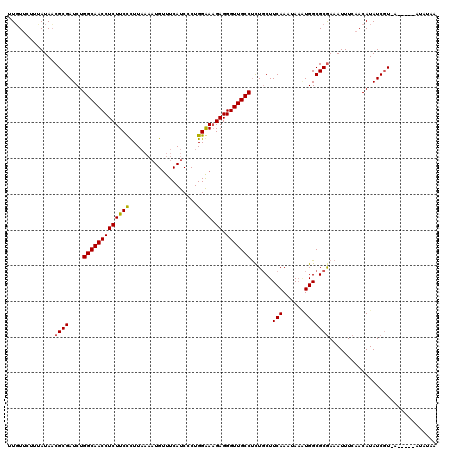
| Location | 4,153,515 – 4,153,635 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -21.64 |
| Energy contribution | -23.20 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4153515 120 - 23771897 UUAUAUUGAAUUUACGAUAUGUUGAAAUUUCGCGCCAUUUUUUUGAAGCAGAGGCAACCCUCUUCCCAGGGAUGAAACAUUUUAAGGGAAGAGGUUGCCAGAUCGCGUUAUGGAGAACAG ..((((((......))))))(((...((..(((((((......))..))...(((((((.(((((((.((((((...))))))..)))))))))))))).....)))..))....))).. ( -34.90) >DroSim_CAF1 3103 112 - 1 UUAUAU----CUGACGAUAUGUUGAAAUUUCGCGCCACUUAUUUGAAGCAGAGGCAACCCUCAUUCCAGGGAUGAAACA----AAGGGAAGAGGUUGCCAGAUCGCGUUAUAAAGUAAAA .(((((----(....)))))).....((..(((((..(......)..))...(((((((.((.((((..(.......).----...))))))))))))).....)))..))......... ( -26.60) >DroYak_CAF1 3289 107 - 1 -------------AUGAUAUGUUCAAAUUUUGCGCCAAAUAAUUGAAGAAAGGGCAACCCUCUUUCCAGGGAUGAAGCAUUUUAAAAGAAGAGGUUGCCAGAUCGCGGUAUAAGGAACAA -------------......(((((.....((((..(((....)))..((...(((((((.(((((...((((((...))))))....))))))))))))...))))))......))))). ( -26.60) >consensus UUAUAU_____U_ACGAUAUGUUGAAAUUUCGCGCCAAUUAUUUGAAGCAGAGGCAACCCUCUUUCCAGGGAUGAAACAUUUUAAGGGAAGAGGUUGCCAGAUCGCGUUAUAAAGAACAA ..............................((((.((......)).......(((((((.(((((((..(((((...)))))...))))))))))))))....))))............. (-21.64 = -23.20 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:33 2006