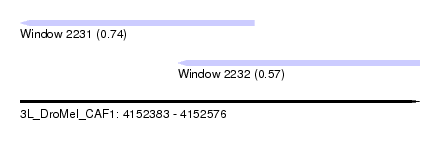
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,152,383 – 4,152,576 |
| Length | 193 |
| Max. P | 0.738616 |

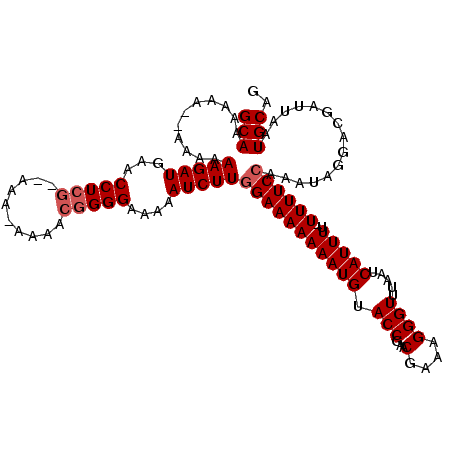
| Location | 4,152,383 – 4,152,496 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.40 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -16.27 |
| Energy contribution | -17.27 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4152383 113 - 23771897 GCAAAAAAAAAAAAAAGAUGAACCUCG--AAA-AAAACGGGGAAAAAUCUUCGAAAAAAAUGUACCGAACGAAAGGGUUUAAUCAUUUUUUUUUCCAAAUAGGACGAUUAAUGCAG (((.......(((((((((((.(((((--...-....)))))..(((((((((............))))(....))))))..)))))))))))(((.....))).......))).. ( -22.80) >DroSim_CAF1 1977 107 - 1 GCAAAA------AAAAGAUGAACCUCG--AAAAAAAACGGGGAAAAAUCUUGGAAAAAAAUGUACCGAACGAAAGGGUUUAAUCAUUUU-UUUUCCAAAUAGGACGAUUAAUGCAG (((...------..(((((...(((((--........)))))....)))))(((((((((((.(((...(....)))).....))))))-)))))................))).. ( -23.50) >DroYak_CAF1 2181 112 - 1 GCAAAAAA--AAAAAAGAUGAACCUGAAGAAA-AAAACGGGGAAAAAUCUUGGAAAAAAAUGUACCGAACGAAAGGGUUUAAUCAUUUU-UUUUCCAAAUAGGACGAUUAAUGCAG (((.....--....(((((...((((......-....)))).....)))))(((((((((((.(((...(....)))).....))))))-)))))................))).. ( -20.60) >consensus GCAAAAAA__AAAAAAGAUGAACCUCG__AAA_AAAACGGGGAAAAAUCUUGGAAAAAAAUGUACCGAACGAAAGGGUUUAAUCAUUUU_UUUUCCAAAUAGGACGAUUAAUGCAG (((...........(((((...(((((..........)))))....)))))(((((((((((.(((...(....)))).....)))))..))))))...............))).. (-16.27 = -17.27 + 1.00)


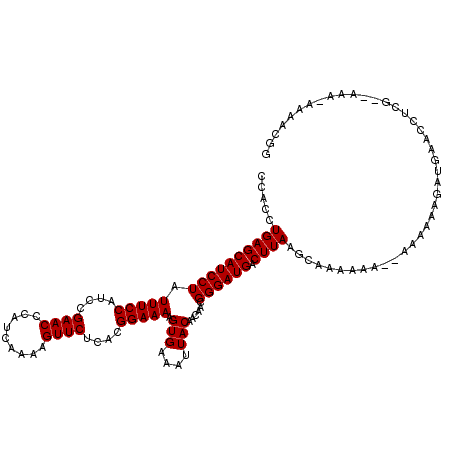
| Location | 4,152,459 – 4,152,576 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.68 |
| Mean single sequence MFE | -17.57 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4152459 117 - 23771897 CCACCUGAGCAUCCUAUUUCCAUCAGAACCCAUCAAAAGUUCUCACGGAAAAGUGAAAUUACAACAACGGGAUGACUUAAGCAAAAAAAAAAAAAAGAUGAACCUCG--AAA-AAAACGG .....((((((((((.(((((...(((((.........)))))...))))).(((....)))......)))))).))))............................--...-....... ( -18.70) >DroSim_CAF1 2052 112 - 1 CCACCUGAGCAUCCUAUUUCCAUCCGAACCCAUCAAAAGUUCUCACGGAAAAGUGAAAUUACAACAACGGGAUGACUUAAGCAAAA------AAAAGAUGAACCUCG--AAAAAAAACGG .....((((((((((.(((((....((((.........))))....))))).(((....)))......)))))).)))).......------...............--........... ( -17.00) >DroYak_CAF1 2256 117 - 1 CCACCUGAGCAUCCUAUUUCCAUACGAACCCAUCAAAAGUUCUCACGGAAAAGUGAAAUUACAACAACGGGAUGACUUAAGCAAAAAA--AAAAAAGAUGAACCUGAAGAAA-AAAACGG .....((((((((((.(((((....((((.........))))....))))).(((....)))......)))))).)))).........--......................-....... ( -17.00) >consensus CCACCUGAGCAUCCUAUUUCCAUCCGAACCCAUCAAAAGUUCUCACGGAAAAGUGAAAUUACAACAACGGGAUGACUUAAGCAAAAAA__AAAAAAGAUGAACCUCG__AAA_AAAACGG .....((((((((((.(((((....((((.........))))....))))).(((....)))......)))))).))))......................................... (-17.00 = -17.00 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:27 2006