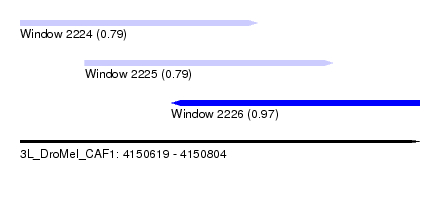
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,150,619 – 4,150,804 |
| Length | 185 |
| Max. P | 0.966880 |

| Location | 4,150,619 – 4,150,729 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -25.55 |
| Energy contribution | -27.66 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4150619 110 + 23771897 CGUCGCGCGAACAAGUUAAGCGACUUCU----------GUUUCUCGGUUAUCCCCACACCCGUCUCUGUUCUUUUUUCUUUUCGGAUCUCGAGAUCGUAAGAUCUUCAGAUCUUAACGGU .(((((...((....))..)))))..((----------(((....((......))............................((((((.((((((....)))))).)))))).))))). ( -27.50) >DroSim_CAF1 260 106 + 1 CGUCGCGCGAACAAGUUAAGCGACUUCU----------GUUUCUCGGUUAUCCCCACACCCGUCUCUUUUCUUUC----UUUGGGAUCUCGAGAUCGUAAGAUCUUCAGAUCUUAACGGU .(((((...((....))..)))))....----------.......((......))....((((............----..((((((((.((((((....)))))).)))))))))))). ( -29.13) >DroYak_CAF1 271 116 + 1 UGUCGCACGAACAAGUUAAGCGACUUCUCGGUUUCUCGGUUUCUCGGCUAUCCCCAUACCCGUCUCUUUUCUUUC----UUUGGGAUCUCGAGAUCGUAAGAUCUUCAGAUCUUAACGGU .(((((((......))...))))).((((((..((((((.....(((.(((....))).))).............----.))))))..))))))((((((((((....)))))).)))). ( -29.51) >consensus CGUCGCGCGAACAAGUUAAGCGACUUCU__________GUUUCUCGGUUAUCCCCACACCCGUCUCUUUUCUUUC____UUUGGGAUCUCGAGAUCGUAAGAUCUUCAGAUCUUAACGGU .(((((((......))...))))).....................((......))....((((..................((((((((.((((((....)))))).)))))))))))). (-25.55 = -27.66 + 2.11)



| Location | 4,150,649 – 4,150,764 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.31 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -25.77 |
| Energy contribution | -28.33 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4150649 115 + 23771897 UUCUCGGUUAUCCCCACACCCGUCUCUGUUCUUUUUUCUUUUCGGAUCUCGAGAUCGUAAGAUCUUCAGAUCUUAACGGUAUGUUUGCAGA-CGCCUAACUGU--GAGCUGG--CAGCCA .....((((..(.(((((...((.(((((........((.((.((((((.((((((....)))))).)))))).)).)).......)))))-.)).....)))--).)..).--.)))). ( -31.76) >DroSim_CAF1 290 111 + 1 UUCUCGGUUAUCCCCACACCCGUCUCUUUUCUUUC----UUUGGGAUCUCGAGAUCGUAAGAUCUUCAGAUCUUAACGGUAUGUUCGCAGA-CGCCUAACUGU--GAGCUGG--CAGCCA .....((((....(((...((((............----..((((((((.((((((....)))))).))))))))))))...((((((((.-.......))))--)))))))--.)))). ( -35.93) >DroYak_CAF1 311 116 + 1 UUCUCGGCUAUCCCCAUACCCGUCUCUUUUCUUUC----UUUGGGAUCUCGAGAUCGUAAGAUCUUCAGAUCUUAACGGUAUGUUUGCAUACCGCCUAACUGUGUGUGCUGGCGCAGCCA .....((((..((.((((((.((............----.(((((((((.((((((....)))))).)))))))))(((((((....)))))))....)).).)))))..))...)))). ( -41.00) >consensus UUCUCGGUUAUCCCCACACCCGUCUCUUUUCUUUC____UUUGGGAUCUCGAGAUCGUAAGAUCUUCAGAUCUUAACGGUAUGUUUGCAGA_CGCCUAACUGU__GAGCUGG__CAGCCA .....((((....(((...((((..................((((((((.((((((....)))))).))))))))))))...((((((((.........))))..)))))))...)))). (-25.77 = -28.33 + 2.56)


| Location | 4,150,689 – 4,150,804 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -34.71 |
| Energy contribution | -35.10 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4150689 115 - 23771897 UCUAGUUAGAAGUGGGUCUCCGGAUCGAUACCAAUUCUCCUGGCUG--CCAGCUC--ACAGUUAGGCG-UCUGCAAACAUACCGUUAAGAUCUGAAGAUCUUACGAUCUCGAGAUCCGAA ............((((((((.((((((.........(.((((((((--.......--.)))))))).)-................(((((((....))))))))))))).)))))))).. ( -37.60) >DroSim_CAF1 326 115 - 1 UCUAGCUAGAAGUGGGUCUCCAGAUCGAUGGCAAUUCUCCUGGCUG--CCAGCUC--ACAGUUAGGCG-UCUGCGAACAUACCGUUAAGAUCUGAAGAUCUUACGAUCUCGAGAUCCCAA ...........(.(((((((.((((((...(((...(.((((((((--.......--.)))))))).)-..)))...........(((((((....))))))))))))).)))))))).. ( -41.30) >DroYak_CAF1 347 120 - 1 ACUAGUUAAAAGUGGGUUUCCGAAUCGAUAGCAAUUCUCCUGGCUGCGCCAGCACACACAGUUAGGCGGUAUGCAAACAUACCGUUAAGAUCUGAAGAUCUUACGAUCUCGAGAUCCCAA ............((((.(..(((((((...........((((((((.(........).))))))))(((((((....))))))).(((((((....))))))))))).)))..).)))). ( -38.80) >consensus UCUAGUUAGAAGUGGGUCUCCGGAUCGAUAGCAAUUCUCCUGGCUG__CCAGCUC__ACAGUUAGGCG_UCUGCAAACAUACCGUUAAGAUCUGAAGAUCUUACGAUCUCGAGAUCCCAA .............(((((((.((((((...(((...(.((((((((.(........).)))))))).)...)))...........(((((((....))))))))))))).)))))))... (-34.71 = -35.10 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:21 2006