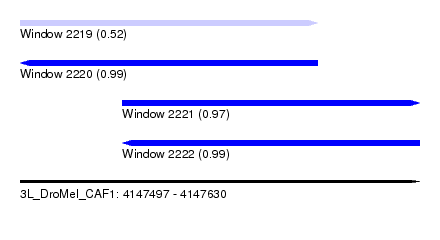
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,147,497 – 4,147,630 |
| Length | 133 |
| Max. P | 0.992341 |

| Location | 4,147,497 – 4,147,596 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -22.19 |
| Energy contribution | -23.75 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4147497 99 + 23771897 GGUGGUGGCGGUGGUGGAGAUGGCGAGUUCAGAGUGCUCUCCGCAAAGGAUAUGAUUUGAGAGAGGAAGUCACCACCUCCUCCGCCUCCAGAGGUCGAA ((.(((((.((.(((((.(((..((....)......(((((..((((........)))).))))))..))).))))).)).))))).)).......... ( -37.90) >DroVir_CAF1 47701 96 + 1 GGCGGUGGAGCCGGUGCUGAGGGUGAGUUGAGCGUGCUCUCCGCAAAGGAUAUGAACUGGGAGAGCAGAUCCCCGCCAGAC---UGUGCAGGCGACGCG ....(((..(((.(..(....((((.(..((...((((((((....((........)).))))))))..)).)))))....---.)..).)))..))). ( -35.50) >DroSim_CAF1 24776 99 + 1 GGUGGCGGCGGUGGUGGAGAUGGCGAGUUCAGAGUGCUCUCCGCAAAGGAUAUGAUUUGAGAGAGGAAGUCACCACCUCCUCCGCCUCCAGAGGACGAA ((.(((((.((.(((((.(((..((....)......(((((..((((........)))).))))))..))).))))).)).))))).)).......... ( -39.80) >DroEre_CAF1 25719 99 + 1 GGUGGCUGUGGUGGUGAAGAUGGGGAGUUCAGAGUGCUCUCCGCAAAGGAUAUGAUUUGCGAGAGGAAGUCACCACCUCCUCCGCCUCCAGAGCUCGAA (((((..(.((((((((....(((((((.......)))))))(((((........))))).........)))))))).)..)))))............. ( -39.60) >DroYak_CAF1 24265 97 + 1 GGUGGCGGCGGUGGUGGAGAUGGCGAGUUCAGAGUGCUCUCCGCAAAGGAAAUGAUUUGGGAAAGGAAGUCCCCGCCUCCUCCGCCUCCAGAGGUCG-- ((.(((((.((.(((((.(((..(((((.......))))(((.((((........)))))))...)..))).))))).)).))))).))........-- ( -41.10) >DroAna_CAF1 26387 87 + 1 GGCGGGGGUGGUGGUGGAGCUGGCGAGUUCAGGGUGCUCUCGGCAAAGGAUAUGAUCUGGGAGAGGAGAUCU---------CCGCCCUGGG---UGGAU ..(.(((((((.......(((((.((((.......))))))))).........(((((........))))).---------))))))).).---..... ( -29.30) >consensus GGUGGCGGCGGUGGUGGAGAUGGCGAGUUCAGAGUGCUCUCCGCAAAGGAUAUGAUUUGGGAGAGGAAGUCACCACCUCCUCCGCCUCCAGAGGUCGAA ((.(((((.((.((((.....((.((((.......)))).))...........(((((........)))))..)))).)).))))).)).......... (-22.19 = -23.75 + 1.56)



| Location | 4,147,497 – 4,147,596 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -18.74 |
| Energy contribution | -20.97 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4147497 99 - 23771897 UUCGACCUCUGGAGGCGGAGGAGGUGGUGACUUCCUCUCUCAAAUCAUAUCCUUUGCGGAGAGCACUCUGAACUCGCCAUCUCCACCACCGCCACCACC .........(((.(((((.(((((((((((.(((((((((((((........)))).))))))......))).)))))))))))....))))).))).. ( -41.20) >DroVir_CAF1 47701 96 - 1 CGCGUCGCCUGCACA---GUCUGGCGGGGAUCUGCUCUCCCAGUUCAUAUCCUUUGCGGAGAGCACGCUCAACUCACCCUCAGCACCGGCUCCACCGCC .(((.....)))...---....((((((((.((((((((((((..........))).)))))))..(((............)))...)).))).))))) ( -31.10) >DroSim_CAF1 24776 99 - 1 UUCGUCCUCUGGAGGCGGAGGAGGUGGUGACUUCCUCUCUCAAAUCAUAUCCUUUGCGGAGAGCACUCUGAACUCGCCAUCUCCACCACCGCCGCCACC .........(((.(((((.(((((((((((.(((((((((((((........)))).))))))......))).)))))))))))....))))).))).. ( -41.70) >DroEre_CAF1 25719 99 - 1 UUCGAGCUCUGGAGGCGGAGGAGGUGGUGACUUCCUCUCGCAAAUCAUAUCCUUUGCGGAGAGCACUCUGAACUCCCCAUCUUCACCACCACAGCCACC ........(((..((.((.((((((((.((.(((((((((((((........))))).)))))......))).)).)))))))).)).)).)))..... ( -32.40) >DroYak_CAF1 24265 97 - 1 --CGACCUCUGGAGGCGGAGGAGGCGGGGACUUCCUUUCCCAAAUCAUUUCCUUUGCGGAGAGCACUCUGAACUCGCCAUCUCCACCACCGCCGCCACC --.......(((.(((((.((((..((.((.(((((((((((((........)))).))))))......))).)).))..))))....))))).))).. ( -35.50) >DroAna_CAF1 26387 87 - 1 AUCCA---CCCAGGGCGG---------AGAUCUCCUCUCCCAGAUCAUAUCCUUUGCCGAGAGCACCCUGAACUCGCCAGCUCCACCACCACCCCCGCC .....---.....(((((---------.(((((........))))).......(((.((((..(.....)..)))).)))..............))))) ( -19.10) >consensus UUCGACCUCUGGAGGCGGAGGAGGUGGUGACUUCCUCUCCCAAAUCAUAUCCUUUGCGGAGAGCACUCUGAACUCGCCAUCUCCACCACCGCCGCCACC .........(((.((.((.(((((.((.((.(((((((((((((........)))).))))))......))).)).)).))))).)).)).)))..... (-18.74 = -20.97 + 2.23)



| Location | 4,147,531 – 4,147,630 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.02 |
| Mean single sequence MFE | -41.12 |
| Consensus MFE | -28.30 |
| Energy contribution | -29.83 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
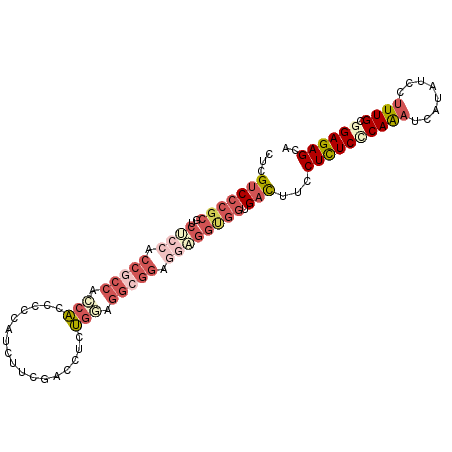
>3L_DroMel_CAF1 4147531 99 + 23771897 UGCUCUCCGCAAAGGAUAUGAUUUGAGAGAGGAAGUCACCACCUCCUCCGCCUCCAGAGGUCGAAGAUGGUGGUGCUGGCGGUGGAGACGCGGGACGAG ..((((((((...........(((....)))...(((.(((((.((..((((.(((...........))).))))..)).))))).))))))))..))) ( -37.00) >DroVir_CAF1 47735 96 + 1 UGCUCUCCGCAAAGGAUAUGAACUGGGAGAGCAGAUCCCCGCCAGAC---UGUGCAGGCGACGCGGCUGCUGGUGCCGGAGCCGGCGCUGCGGGACGCG ((((((((....((........)).))))))))....(((((.....---...((((.((...)).)))).(((((((....))))))))))))..... ( -44.50) >DroSim_CAF1 24810 99 + 1 UGCUCUCCGCAAAGGAUAUGAUUUGAGAGAGGAAGUCACCACCUCCUCCGCCUCCAGAGGACGAAGAUGGGGGUGGUGGCGGUGGGGACGCGGGACGAG ..((((((((...........(((....)))...(((.(((((.((.(((((((((...........))))))))).)).))))).))))))))..))) ( -44.20) >DroEre_CAF1 25753 99 + 1 UGCUCUCCGCAAAGGAUAUGAUUUGCGAGAGGAAGUCACCACCUCCUCCGCCUCCAGAGCUCGAAGAUGGGGGUGGGGGCGGUGGAGAUGCGGGACGAG ..(((((((((.......((((((.(....).))))))(((((.((((((((((((...(.....).)))))))))))).)))))...))))))..))) ( -51.20) >DroYak_CAF1 24299 96 + 1 UGCUCUCCGCAAAGGAAAUGAUUUGGGAAAGGAAGUCCCCGCCUCCUCCGCCUCCAGAGGUCG---AUGGGGGUGGUGGUGGGGGAGACGCGGGACGAG ..((((((.((((........)))))))......(((((((.((((((((((.(((.......---.......))).)))))))))).)).)))))))) ( -42.14) >DroAna_CAF1 26421 87 + 1 UGCUCUCGGCAAAGGAUAUGAUCUGGGAGAGGAGAUCU---------CCGCCCUGGG---UGGAUGAGGAGGGCGGCGGCGGUGGAGAGGCUGGGCGGG ..(((((.((.........(((((........))))).---------(((((((...---.........))))))).....)).))))).......... ( -27.70) >consensus UGCUCUCCGCAAAGGAUAUGAUUUGGGAGAGGAAGUCACCACCUCCUCCGCCUCCAGAGGUCGAAGAUGGGGGUGGUGGCGGUGGAGACGCGGGACGAG .(.((.((((.........(((((........))))).(((((.((.(((((((((...........))))))))).)).)))))....)))))))... (-28.30 = -29.83 + 1.53)


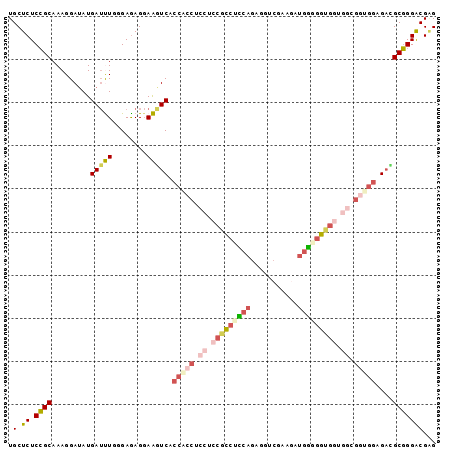
| Location | 4,147,531 – 4,147,630 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.02 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -21.75 |
| Energy contribution | -24.28 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4147531 99 - 23771897 CUCGUCCCGCGUCUCCACCGCCAGCACCACCAUCUUCGACCUCUGGAGGCGGAGGAGGUGGUGACUUCCUCUCUCAAAUCAUAUCCUUUGCGGAGAGCA ........((.(((((......(((((((((..(((((.(((....))))))))..))))))).))........((((........)))).))))))). ( -35.60) >DroVir_CAF1 47735 96 - 1 CGCGUCCCGCAGCGCCGGCUCCGGCACCAGCAGCCGCGUCGCCUGCACA---GUCUGGCGGGGAUCUGCUCUCCCAGUUCAUAUCCUUUGCGGAGAGCA ....((((((.((((((....))))....((((.((...)).))))...---))...))))))....((((((((((..........))).))))))). ( -40.30) >DroSim_CAF1 24810 99 - 1 CUCGUCCCGCGUCCCCACCGCCACCACCCCCAUCUUCGUCCUCUGGAGGCGGAGGAGGUGGUGACUUCCUCUCUCAAAUCAUAUCCUUUGCGGAGAGCA ...(((((((.(((...(((((.(((.................))).))))).))).)))).)))...((((((((((........)))).)))))).. ( -35.43) >DroEre_CAF1 25753 99 - 1 CUCGUCCCGCAUCUCCACCGCCCCCACCCCCAUCUUCGAGCUCUGGAGGCGGAGGAGGUGGUGACUUCCUCUCGCAAAUCAUAUCCUUUGCGGAGAGCA ...(((.(.(((((((.(((((.(((.................))).))))).)))))))).)))...((((((((((........))))).))))).. ( -38.43) >DroYak_CAF1 24299 96 - 1 CUCGUCCCGCGUCUCCCCCACCACCACCCCCAU---CGACCUCUGGAGGCGGAGGAGGCGGGGACUUCCUUUCCCAAAUCAUUUCCUUUGCGGAGAGCA ...(((((.(((((((.((.((.(((.......---.......))).)).)).))))))))))))...((((((((((........)))).)))))).. ( -39.94) >DroAna_CAF1 26421 87 - 1 CCCGCCCAGCCUCUCCACCGCCGCCGCCCUCCUCAUCCA---CCCAGGGCGG---------AGAUCUCCUCUCCCAGAUCAUAUCCUUUGCCGAGAGCA ........((.((((........(((((((.........---...)))))))---------.(((((........)))))............)))))). ( -23.90) >consensus CUCGUCCCGCGUCUCCACCGCCACCACCCCCAUCUUCGACCUCUGGAGGCGGAGGAGGUGGUGACUUCCUCUCCCAAAUCAUAUCCUUUGCGGAGAGCA ...(((((((..((((.(((((.(((.................))).))))).)))))))).)))...((((((((((........)))).)))))).. (-21.75 = -24.28 + 2.53)

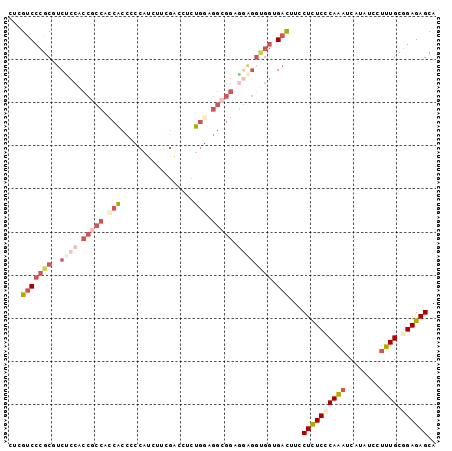
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:17 2006