
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,134,897 – 4,135,011 |
| Length | 114 |
| Max. P | 0.543294 |

| Location | 4,134,897 – 4,135,011 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -44.39 |
| Consensus MFE | -32.93 |
| Energy contribution | -33.13 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513194 |
| Prediction | RNA |
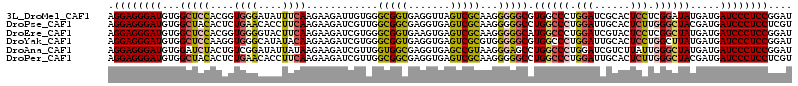
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4134897 114 + 23771897 AGGAGGGAUGUGGCUCCACGGUGGGAUAUUUCAAGAAGAUUGUGGGCGGUGAGGUUAGUCGCAAGGGGGCGUGGCCCUGGAUCGCACUCCUCGGAUAUGAUGAUCCCUCCGGAU .((((((((...((.(((((((..((....))......)))))))))..(((((..(((((....(((((...)))))....)).)))))))).........)))))))).... ( -40.40) >DroPse_CAF1 11058 114 + 1 AGGAGGGAUGUGGCUACACUCUGAACACCUUCAAGAAGAUCGUUGGCGGCGAGGUGAGUCGCAAGGGGGCCUGGCCCUGGAUUGCACUCUUGGGCUACGAUGAUCCCUCCUCGU (((((((((((((((((.(((((((....))))....(.(((....)))))))))((((.((((.(((((...)))))...))))))))...))))))....)))))))))... ( -43.30) >DroEre_CAF1 13099 114 + 1 AGGAGGGAUGUGGCUCCACGGUGGGGUACUUCAAGAAGAUCGUGGGCGGUGAAGUGAGUCGCAAGGGGGCAUGGCCCUGGAUCGUACUCCUCGGCUAUGAUGAUCCCUCCGGAU .((((((((...(((((...((((..(((((((......(((....))))))))))..))))...)))))((((((..(((......)))..))))))....)))))))).... ( -49.80) >DroYak_CAF1 11393 114 + 1 AGGAGGGAUGUGGCUCCAAGGUGGGCAUAUACAAGAAGAUCGUGGGCGGUGAGGUGAGUCGCGUGGGGGCGUGGCCCUGGAUUGCACUCCUGGCUUAUGAUGAUCCCUCCGGAU .((((((((...((((......))))............(((((((((.....((.((((.((.(.(((((...))))).)...)))))))).))))))))).)))))))).... ( -44.70) >DroAna_CAF1 10756 114 + 1 AGGAGGGAUGUGGAUCUACUGUCGGAUAUUAUAAGAAGAUCGUUGGUGGCGAGGUGAGCCGUAAGGGAGCCUGGCCCUGGAUCGUCUUAUUGGGCUAUGAUGAUCCCUCCGGAU .(((((((((((....))).((((......((((((.((((...((.(((.((((...((....))..)))).))))).)))).)))))).......)))).)))))))).... ( -44.82) >DroPer_CAF1 10209 114 + 1 AGGAGGGAUGUGGCUACACUCUGAACACCUUCAAGAAGAUCGUUGGCGGCGAGGUGAGUCGCAAGGGGGCCUGGCCCUGGAUUGCACUCUUGGGCUACGAUGAUCCCUCCUCGU (((((((((((((((((.(((((((....))))....(.(((....)))))))))((((.((((.(((((...)))))...))))))))...))))))....)))))))))... ( -43.30) >consensus AGGAGGGAUGUGGCUCCACGGUGGGCAACUUCAAGAAGAUCGUGGGCGGCGAGGUGAGUCGCAAGGGGGCCUGGCCCUGGAUCGCACUCCUGGGCUAUGAUGAUCCCUCCGGAU .((((((((...(((((....((((....))))............(((((.......)))))...))))).((((((.(((......))).)))))).....)))))))).... (-32.93 = -33.13 + 0.20)
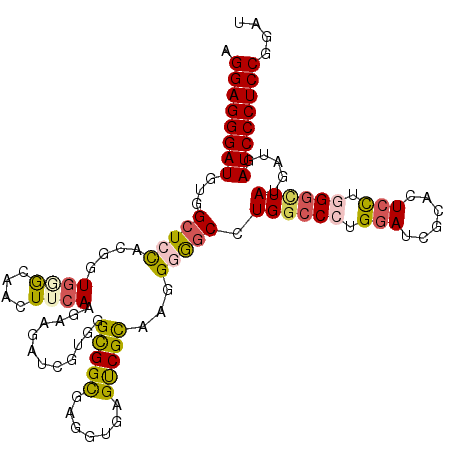
| Location | 4,134,897 – 4,135,011 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -22.70 |
| Energy contribution | -24.23 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4134897 114 - 23771897 AUCCGGAGGGAUCAUCAUAUCCGAGGAGUGCGAUCCAGGGCCACGCCCCCUUGCGACUAACCUCACCGCCCACAAUCUUCUUGAAAUAUCCCACCGUGGAGCCACAUCCCUCCU ....((((((((..........(((((((((((....((((...))))..)))).)))..))))...((((((......................)))).))...)))))))). ( -36.25) >DroPse_CAF1 11058 114 - 1 ACGAGGAGGGAUCAUCGUAGCCCAAGAGUGCAAUCCAGGGCCAGGCCCCCUUGCGACUCACCUCGCCGCCAACGAUCUUCUUGAAGGUGUUCAGAGUGUAGCCACAUCCCUCCU ...(((((((((....((.((....((((((((....((((...))))..)))).))))(((((..((((..(((.....)))..))))....))).)).)).))))))))))) ( -39.50) >DroEre_CAF1 13099 114 - 1 AUCCGGAGGGAUCAUCAUAGCCGAGGAGUACGAUCCAGGGCCAUGCCCCCUUGCGACUCACUUCACCGCCCACGAUCUUCUUGAAGUACCCCACCGUGGAGCCACAUCCCUCCU ....((((((((..........(((((((.((.....((((...)))).....)))))).)))....(((((((....................))))).))...)))))))). ( -36.35) >DroYak_CAF1 11393 114 - 1 AUCCGGAGGGAUCAUCAUAAGCCAGGAGUGCAAUCCAGGGCCACGCCCCCACGCGACUCACCUCACCGCCCACGAUCUUCUUGUAUAUGCCCACCUUGGAGCCACAUCCCUCCU ....((((((((.........(((((.(((.......((((...((......))((......))...))))((((.....)))).......))))))))......)))))))). ( -31.96) >DroAna_CAF1 10756 114 - 1 AUCCGGAGGGAUCAUCAUAGCCCAAUAAGACGAUCCAGGGCCAGGCUCCCUUACGGCUCACCUCGCCACCAACGAUCUUCUUAUAAUAUCCGACAGUAGAUCCACAUCCCUCCU ....((((((((............((((((.((((...(((.(((...((....))....))).)))......)))).))))))...........((......)))))))))). ( -31.40) >DroPer_CAF1 10209 114 - 1 ACGAGGAGGGAUCAUCGUAGCCCAAGAGUGCAAUCCAGGGCCAGGCCCCCUUGCGACUCACCUCGCCGCCAACGAUCUUCUUGAAGGUGUUCAGAGUGUAGCCACAUCCCUCCU ...(((((((((....((.((....((((((((....((((...))))..)))).))))(((((..((((..(((.....)))..))))....))).)).)).))))))))))) ( -39.50) >consensus AUCCGGAGGGAUCAUCAUAGCCCAAGAGUGCAAUCCAGGGCCAGGCCCCCUUGCGACUCACCUCACCGCCAACGAUCUUCUUGAAGUAGCCCACAGUGGAGCCACAUCCCUCCU ....((((((((.............((((((((....((((...))))..)))).))))....................................(((....))))))))))). (-22.70 = -24.23 + 1.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:13 2006