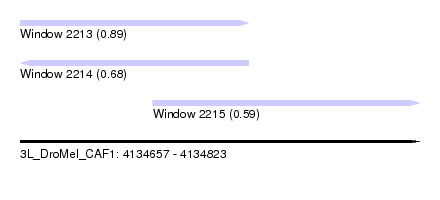
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,134,657 – 4,134,823 |
| Length | 166 |
| Max. P | 0.893351 |

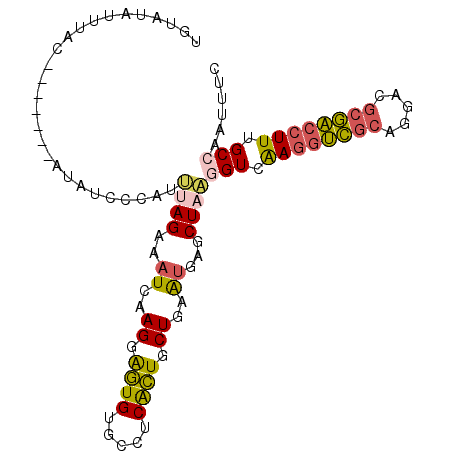
| Location | 4,134,657 – 4,134,752 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -26.11 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.37 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4134657 95 + 23771897 UGUAUAUUUAC-------UUAUCCCAUUUAGAAAUCAAGGAGUGUGCCUCACUGCUGAAUGAGCUUAGGUCAAGGUCGCAGGACGCGACCUUUGCCAAUUUC .(((....)))-------......(((((((......(((......))).....)))))))......(((.((((((((.....)))))))).)))...... ( -27.30) >DroVir_CAF1 14619 98 + 1 U--ACA--GAGCUCAUACUAAUCCCUUGCAGAAAUCAAGAGCUGUGCGCCGGUGCUAAGCGAACUCCAGUUGAAGCGUAACGACCCCGACUUUGCCAAGCAA .--.((--.(((((........................))))).))......((((..(((((.((..((((........))))...)).)))))..)))). ( -19.46) >DroSec_CAF1 10921 95 + 1 AAUAUAUUUAC-------AUAUCCCAUUUAGAAAUCAAGGAGUGUGCCUCACUGCUGAAUGAGCUAAGGUCAAGGUCGCAGGACGCGACCUUUGCAAAUUUC ...........-------........(((((...(((((.((((.....)))).))...))).)))))((.((((((((.....)))))))).))....... ( -25.10) >DroSim_CAF1 11983 95 + 1 AAUGUAUUUAC-------AUAUCCCAUUUAGAAAUCAAGGAGUGUGCCUCACUGCUGAAUGAGCUAAGGUCAAGGUCGCAGGACGCGACCUUUGCCAAUUUC .((((....))-------)).......((((...(((((.((((.....)))).))...))).))))(((.((((((((.....)))))))).)))...... ( -28.70) >DroEre_CAF1 12860 94 + 1 UGAAUAUUUAC-------CUAUCCCAU-UAGAAAUCAAGGAGUGUGCCUCGCUGCUGAAUGAGCUAAGGUCAAGGUCGCAGGACGCGACCUUUGCCAAUUUC ...........-------........(-(((...(((((.((((.....)))).))...))).))))(((.((((((((.....)))))))).)))...... ( -27.60) >DroYak_CAF1 11144 95 + 1 UGAAUAUUAAC-------CUAUUCCAUUUAGAAAUCAAGGAGUGCGCCACGCUGCUGAAUGAGCUAAGGUCAAGGUCGCAGGACGCGACCUUUGCCAACUUC .(((((.....-------.)))))...((((...(((((.((((.....)))).))...))).))))(((.((((((((.....)))))))).)))...... ( -28.50) >consensus UGUAUAUUUAC_______AUAUCCCAUUUAGAAAUCAAGGAGUGUGCCUCACUGCUGAAUGAGCUAAGGUCAAGGUCGCAGGACGCGACCUUUGCCAAUUUC ...........................((((..((..((.((((.....)))).))..))...))))(((.((((((((.....)))))))).)))...... (-21.42 = -21.37 + -0.05)

| Location | 4,134,657 – 4,134,752 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -17.83 |
| Energy contribution | -17.83 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4134657 95 - 23771897 GAAAUUGGCAAAGGUCGCGUCCUGCGACCUUGACCUAAGCUCAUUCAGCAGUGAGGCACACUCCUUGAUUUCUAAAUGGGAUAA-------GUAAAUAUACA ......((..((((((((.....))))))))..))....((((((.((.(((((((......)))).))).)).))))))....-------(((....))). ( -25.30) >DroVir_CAF1 14619 98 - 1 UUGCUUGGCAAAGUCGGGGUCGUUACGCUUCAACUGGAGUUCGCUUAGCACCGGCGCACAGCUCUUGAUUUCUGCAAGGGAUUAGUAUGAGCUC--UGU--A ((((...)))).(((((.((.((...(((((....)))))..))...)).)))))(((.((((((((((..(.....)..)))))...))))).--)))--. ( -27.00) >DroSec_CAF1 10921 95 - 1 GAAAUUUGCAAAGGUCGCGUCCUGCGACCUUGACCUUAGCUCAUUCAGCAGUGAGGCACACUCCUUGAUUUCUAAAUGGGAUAU-------GUAAAUAUAUU ...(((((((((((((((.....))))))))........((((((.((.(((((((......)))).))).)).))))))...)-------))))))..... ( -27.00) >DroSim_CAF1 11983 95 - 1 GAAAUUGGCAAAGGUCGCGUCCUGCGACCUUGACCUUAGCUCAUUCAGCAGUGAGGCACACUCCUUGAUUUCUAAAUGGGAUAU-------GUAAAUACAUU ......((..((((((((.....))))))))..))....((((((.((.(((((((......)))).))).)).))))))..((-------((....)))). ( -25.30) >DroEre_CAF1 12860 94 - 1 GAAAUUGGCAAAGGUCGCGUCCUGCGACCUUGACCUUAGCUCAUUCAGCAGCGAGGCACACUCCUUGAUUUCUA-AUGGGAUAG-------GUAAAUAUUCA (((.......((((((((.....)))))))).((((...((((((.((...(((((......)))))....)))-)))))..))-------)).....))). ( -29.60) >DroYak_CAF1 11144 95 - 1 GAAGUUGGCAAAGGUCGCGUCCUGCGACCUUGACCUUAGCUCAUUCAGCAGCGUGGCGCACUCCUUGAUUUCUAAAUGGAAUAG-------GUUAAUAUUCA ((((((((..((((((((.....))))))))..))...(((.....))).(((...))).......))))))......(((((.-------.....))))). ( -26.10) >consensus GAAAUUGGCAAAGGUCGCGUCCUGCGACCUUGACCUUAGCUCAUUCAGCAGCGAGGCACACUCCUUGAUUUCUAAAUGGGAUAA_______GUAAAUAUACA ......((..((((((((.....))))))))..))....((((((.(((((.(((.....))).)))....)).))))))...................... (-17.83 = -17.83 + 0.01)

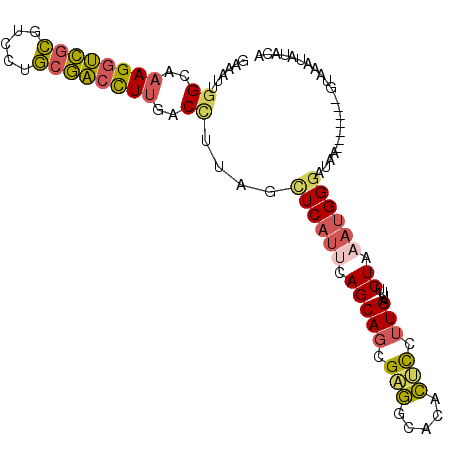

| Location | 4,134,712 – 4,134,823 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.61 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -22.46 |
| Energy contribution | -24.61 |
| Covariance contribution | 2.15 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4134712 111 + 23771897 GCUUAGGUCAAGGUCGCAGGACGCGACCUUUGCCAAUUUCCUGCGUGCCUCGAACGCGGUUUGCCAAAACAAGGGCACCCAGGUGUGCUGCCCCACUGGCCAGGG---CAUUAC .....(((.((((((((.....)))))))).))).....((((.((((((.......((....)).......)))))).))))(((.(((((.....)).))).)---)).... ( -43.94) >DroVir_CAF1 14677 111 + 1 ACUCCAGUUGAAGCGUAACGACCCCGACUUUGCCAAGCAAUUGCAGGCCUCCAAUUUGGUAUGCGGCAGGGUUGGCACCCAGGUCUGUUGUCCCACGGGCCAAAC---GGUGAC ......((((........)))).(((((((((((((....))(((.(((........))).))))))))))))))((((..(((((((......)))))))....---)))).. ( -39.40) >DroSec_CAF1 10976 111 + 1 GCUAAGGUCAAGGUCGCAGGACGCGACCUUUGCAAAUUUCCUGCGUGCCUCGAACGCGGUUUGCCAGAACAAGGGCACCCAGGUGUGCUGUCCCACUGGCCAGGG---CAUCAC (((..((((((((((((.....)))))))).(((.....((((.((((((.......((....)).......)))))).))))..))).........))))..))---)..... ( -43.64) >DroSim_CAF1 12038 111 + 1 GCUAAGGUCAAGGUCGCAGGACGCGACCUUUGCCAAUUUCCUGCGUGCCUCGAACGCGGUCUGCCAGAACAAGGGCACCCAGGUGUGCUGUCCCACUGGCCAGGG---CAUCAC (((..(((.((((((((.....)))))))).)))......((((((.......)))))).(((((((....((.((((....)))).))......)))).)))))---)..... ( -46.10) >DroYak_CAF1 11199 114 + 1 GCUAAGGUCAAGGUCGCAGGACGCGACCUUUGCCAACUUCCUGCGUGCCUCCAACACCAUUUGCCAGAACAGGGGCACCCAGGUGUGCUGCCCCACCGCACAGGGCAACAACGC ((...(((.((((((((.....)))))))).))).((..((((.(((((((....................))))))).)))).))))(((((.........)))))....... ( -43.75) >DroPer_CAF1 10033 111 + 1 ACUGAUUGCCAAGCAGAAGGAUUCCACCUUCGUGAGCUUCCUGCAGGCCUCGCAGCGCAUCUGCGGCAACGUAGGCUCCACGGUCUGCUGCCCCAAUGGCCAGAC---GGCGAC .....(((((.....(((((......)))))((((((...((((.......)))).....(((((....)))))))).))).(((((..(((.....))))))))---))))). ( -42.20) >consensus GCUAAGGUCAAGGUCGCAGGACGCGACCUUUGCCAAUUUCCUGCGUGCCUCGAACGCGGUUUGCCAGAACAAGGGCACCCAGGUGUGCUGCCCCACUGGCCAGGG___CAUCAC .....(((.((((((((.....)))))))).))).....((((.((((((.......((....)).......)))))).))))....(((.((....)).)))........... (-22.46 = -24.61 + 2.15)

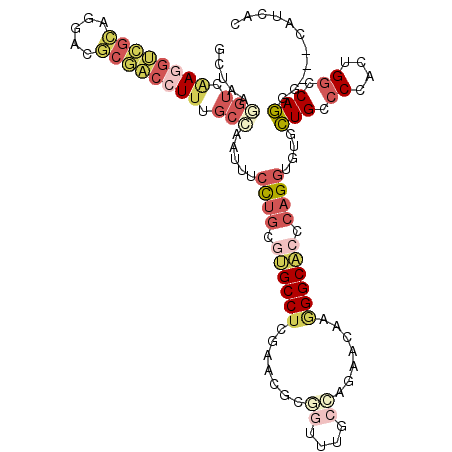
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:11 2006