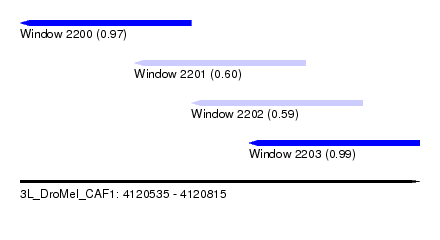
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,120,535 – 4,120,815 |
| Length | 280 |
| Max. P | 0.986113 |

| Location | 4,120,535 – 4,120,655 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -38.26 |
| Energy contribution | -39.10 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4120535 120 - 23771897 GACCAAUGCGGUUCGCAAUUGGGAGGUCCUUGUUGGCUCUUCGCACAUCCUGUCGCCCGAAAUCUUCCUCAGCGAUUUGGGCAGUCUCUCGAAGGAGGACUAAGGAACUUAUUGGGUCAU ((((..(((((...(((((.(((....))).))).))...)))))..((((...((((.(((((.........)))))))))((((.(((....))))))).))))........)))).. ( -43.20) >DroSec_CAF1 2154 120 - 1 GACCAAUGCGGUUCGCAAUUGGGAGGUCCUUGUGGGCUCUUCGCACAUCCUGUCGCCCGAAAUCUUCCUCAGCGAUUUGGGCAGUCUCUCGAAGGAGGACUAAGGAACUUAUUCGGUCAU ((((..(((((((((((...(((....))))))))))....))))..((((...((((.(((((.........)))))))))((((.(((....))))))).))))........)))).. ( -44.40) >DroSim_CAF1 2450 120 - 1 GACCAAUGCUGUUCGCAAUUGGGAGGUCCUUGUGGGCUCUUCGCACAUCCUGUCGCCCGAAAUCUUCCUCAGCGAUUUGGGCAGUCUCUCGAAGGAGGACUAAGGAACUUAUUCGGUCAU ((((..(((((....))....((((((((....)))).)))))))..((((...((((.(((((.........)))))))))((((.(((....))))))).))))........)))).. ( -43.20) >DroEre_CAF1 2350 120 - 1 GACCAAUGCGGUUCGCAAUUGGGAGGUCCUUGUGGGCUCUUCGCACAUCCUGUCGCCCGAAAUCUUCCUCAGCGAUUUGGGCAGUCUCUCAAAGGAGGACUAAAGAACUUAUUGGGUCGU ((((((((.(((((((...((((((((((....)))).)))).))...((.(((((..((........)).)))))..))))((((.(((....)))))))...))))))))).)))).. ( -39.80) >DroYak_CAF1 2800 120 - 1 GACCAAUGCGGUUCGCAAUUGGGAAGUCCUUGUGGGCUCCUCGCACAUCCUGUCGCCCGAAAUAUUCCUCAGCGAUUUGGGUAGUCUCUCGAAGGAGGACUAAAGAGCUAAUUGGGUCAU (((((((..(((((((....((((.((((....)))))))).))..((((.(((((..((........)).)))))..))))((((.(((....)))))))...))))).))).)))).. ( -43.00) >consensus GACCAAUGCGGUUCGCAAUUGGGAGGUCCUUGUGGGCUCUUCGCACAUCCUGUCGCCCGAAAUCUUCCUCAGCGAUUUGGGCAGUCUCUCGAAGGAGGACUAAGGAACUUAUUGGGUCAU ((((..(((((((((((...(((....))))))))))....))))..((((...((((.(((((.........)))))))))((((.(((....))))))).))))........)))).. (-38.26 = -39.10 + 0.84)


| Location | 4,120,615 – 4,120,735 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -47.30 |
| Consensus MFE | -46.48 |
| Energy contribution | -46.36 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4120615 120 - 23771897 CAGGUAAAGAACGUGCCCAGGCUUAUUGUCUUCAUCGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGUGACCAAUGCGGUUCGCAAUUGGGAGGUCCUUGUUGGCUCU .......(((..(..((.((((.....)))).((((.((((((((....)).)))))).)))).))..)(((.((((((...((((((((...))).))))).....)))))).)))))) ( -44.80) >DroSec_CAF1 2234 120 - 1 CAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUCGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGUGACCAAUGCGGUUCGCAAUUGGGAGGUCCUUGUGGGCUCU .......(((..(..((.((((.....)))).((((.((((((((....)).)))))).)))).))..)((((((((((...((((((((...))).))))).....))))))))))))) ( -49.00) >DroSim_CAF1 2530 120 - 1 CAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUCGUGGGUGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGUGACCAAUGCUGUUCGCAAUUGGGAGGUCCUUGUGGGCUCU .......(((..(..((.((((.....)))).((((.((((..((....))...)))).)))).))..)((((((((((...(((((((.....)).))))).....))))))))))))) ( -44.90) >DroEre_CAF1 2430 120 - 1 CAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUUGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGUGACCAAUGCGGUUCGCAAUUGGGAGGUCCUUGUGGGCUCU .......(((..(..((.((((.....)))).((((.((((((((....)).)))))).)))).))..)((((((((((...((((((((...))).))))).....))))))))))))) ( -47.30) >DroYak_CAF1 2880 120 - 1 CAGGUGAAGAAUGUGCCCAGGCUCAUUGUCUUCAUCGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGUGACCAAUGCGGUUCGCAAUUGGGAAGUCCUUGUGGGCUCC ..(((((((((((.((....)).)))..)))))))).((((((((....)).))))))......((...((((((((((...((((((((...))).))))).....)))))))))).)) ( -50.50) >consensus CAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUCGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGUGACCAAUGCGGUUCGCAAUUGGGAGGUCCUUGUGGGCUCU ............(..((.((((.....)))).((((.((((((((....)).)))))).)))).))..)((((((((((...(((((((.....)).))))).....))))))))))... (-46.48 = -46.36 + -0.12)


| Location | 4,120,655 – 4,120,775 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -49.62 |
| Consensus MFE | -49.20 |
| Energy contribution | -48.76 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4120655 120 - 23771897 UGCCCGCUACGGUCACUGGCACAAGGACAAGGGACAGGCGCAGGUAAAGAACGUGCCCAGGCUUAUUGUCUUCAUCGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGU ((((.((....))....))))..((((((((((...((.(((.((.....)).)))))...))).)))))))..(((((((((((....))....(((.....)))..)))))))))... ( -42.60) >DroSec_CAF1 2274 120 - 1 UGCCCGCUACGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUCGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGU ((((.((....))....))))..((((((((((((.((.(((.((.....)).))))).))))).)))))))..(((((((((((....))....(((.....)))..)))))))))... ( -52.50) >DroSim_CAF1 2570 120 - 1 UGCCCGCUACGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUCGUGGGUGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGU ((((.((....))....))))..((((((((((((.((.(((.((.....)).))))).))))).)))))))..(((((((((((....))....(((.....)))..)))))))))... ( -50.60) >DroEre_CAF1 2470 120 - 1 UGCCCGCUAUGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUUGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGU .((((((.(((((.....))...((((((((((((.((.(((.((.....)).))))).))))).)))))))))).))))))((((((.((((.......))))..))))))........ ( -50.80) >DroYak_CAF1 2920 120 - 1 UGCCCGCUAUGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAAUGUGCCCAGGCUCAUUGUCUUCAUCGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGU .(((((.((.((((((((.....((((((((((((.((((((.........))))))..))))).)))))))((((.((((((((....)).)))))).))))))))).))))))).))) ( -51.60) >consensus UGCCCGCUACGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUCGUGGGCGGCGUGAGUAUGUCCGAGAUGCGGUGCGCCUACGAGGU .(((((.((.((((((((.....((((((((((((.((.(((.((.....)).))))).))))).)))))))((((.((((((((....)).)))))).))))))))).))))))).))) (-49.20 = -48.76 + -0.44)



| Location | 4,120,695 – 4,120,815 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -43.86 |
| Energy contribution | -43.50 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4120695 120 - 23771897 AGGGCAGGGCCCAAAAUACCAAUUAUCAUGCCCCAACCAGUGCCCGCUACGGUCACUGGCACAAGGACAAGGGACAGGCGCAGGUAAAGAACGUGCCCAGGCUUAUUGUCUUCAUCGUGG .(((((.((.........))........)))))...((((((.(((...))).))))))(((.((((((((((...((.(((.((.....)).)))))...))).)))))))....))). ( -36.50) >DroSec_CAF1 2314 120 - 1 AGGGCAGGGCCCAGAAUACCAAUUAUCAUGCCCCAACCAGUGCCCGCUACGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUCGUGG .(((((.((.........))........)))))...((((((.(((...))).))))))(((.((((((((((((.((.(((.((.....)).))))).))))).)))))))....))). ( -46.40) >DroSim_CAF1 2610 120 - 1 AGGGCAGGGCCCAGAAUACCAAUUAUCAUGCCCCAACCAGUGCCCGCUACGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUCGUGG .(((((.((.........))........)))))...((((((.(((...))).))))))(((.((((((((((((.((.(((.((.....)).))))).))))).)))))))....))). ( -46.40) >DroEre_CAF1 2510 120 - 1 AGGGCAGGGCCCAGAACACCAAUUAUCAUGCCCCAACCAGUGCCCGCUAUGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUUGUGG .(((((.((.........))........)))))...((((((.((.....)).))))))((((((((((((((((.((.(((.((.....)).))))).))))).))))))...))))). ( -47.40) >DroYak_CAF1 2960 120 - 1 AGGGCAGGGCACAGAAUACCAAUUAUCAUGCCCCAACCAGUGCCCGCUAUGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAAUGUGCCCAGGCUCAUUGUCUUCAUCGUGG .(((((.((.........))........)))))...((((((.((.....)).))))))(((.((((((((((((.((((((.........))))))..))))).)))))))....))). ( -44.30) >consensus AGGGCAGGGCCCAGAAUACCAAUUAUCAUGCCCCAACCAGUGCCCGCUACGGUCACUGGCACAAGGACAAGGGCCAGGCGCAGGUGAAGAACGUGCCCAGGCUCAUUGUCUUCAUCGUGG .(((((.((.........))........)))))...((((((.(((...))).))))))(((.((((((((((((.((.(((.((.....)).))))).))))).)))))))....))). (-43.86 = -43.50 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:59 2006