
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,116,831 – 4,116,934 |
| Length | 103 |
| Max. P | 0.715270 |

| Location | 4,116,831 – 4,116,934 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -26.11 |
| Consensus MFE | -14.86 |
| Energy contribution | -16.82 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4116831 103 + 23771897 UUUAGAAACUACAGUG-AUUUAAAGCGUGUCACUGGGUGAGAAUAGAUAAGAAGAUUUUCUUAUUAGAAAAACAGCUAUGAAAGACUUGCA-AUAGACCAGUGAC-------------- ((((((.((....)).-.))))))....(((((((((..((....((((((((....)))))))).......((....)).....))..).-.....))))))))-------------- ( -20.00) >DroSec_CAF1 18358 114 + 1 UUAAGAAACUACAGUG-AUUUAAAGCGUGUCACUGGUUGAGAAUAGAUAA---GAUUUUCAUAUUAAAAAAACAGCAAUGAAAGACUUGCA-AUAGACCAGUGACAGUGGUAUUCCGGG ....((((((((.((.-.......)).(((((((((((((((((......---.))))))..............((((........)))).-...)))))))))))))))).))).... ( -29.90) >DroSim_CAF1 18754 117 + 1 UUAAGAAACUACAGUG-AUUUAAAGCGUGUCACUGGUUGAGAAUAGAUAAGAAGAUUUUCAUAUUAGAAAAACAGCAAUGAAAGACUUGCA-AUAGACCAGUGACAGUGGUAUUCCGGG ....((((((((.((.-.......)).(((((((((((((((((..........))))))..............((((........)))).-...)))))))))))))))).))).... ( -29.30) >DroEre_CAF1 18602 117 + 1 UUAAGUAACUACUGGGGAUACAAA-UGUGUCAAUGGUUGAGAAUAGAUAAGAAGAUUUUCCUAUUAGAAAAACCGUACUGAAAAACUAGCA-AUAUACCAGUGUCAGUGGUAUUCUGGG .......((((((((.(((((...-.))))).((((((...(((((..(((....)))..))))).....))))))((((...........-......)))).))))))))........ ( -22.63) >DroYak_CAF1 18825 117 + 1 UUAAA-AACUACUGGAGACAUAAAGUGUGUCACUGGUUGAU-AUAGAUAAGAUGAUUUUCCUAUUAGAAAAGCGGUACUGAAAAACUUGCAAAUAGACCAGUGACAGUGGUAUUCCGGG .....-.(((((((....)).......(((((((((((...-.............(((((......)))))(((((........)).))).....))))))))))))))))........ ( -28.70) >consensus UUAAGAAACUACAGUG_AUUUAAAGCGUGUCACUGGUUGAGAAUAGAUAAGAAGAUUUUCAUAUUAGAAAAACAGCAAUGAAAGACUUGCA_AUAGACCAGUGACAGUGGUAUUCCGGG ...........................(((((((((((((((((..........))))))..............((((........)))).....)))))))))))............. (-14.86 = -16.82 + 1.96)

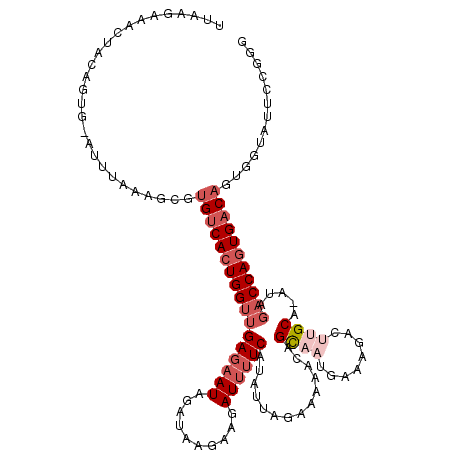
| Location | 4,116,831 – 4,116,934 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -20.09 |
| Consensus MFE | -14.12 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4116831 103 - 23771897 --------------GUCACUGGUCUAU-UGCAAGUCUUUCAUAGCUGUUUUUCUAAUAAGAAAAUCUUCUUAUCUAUUCUCACCCAGUGACACGCUUUAAAU-CACUGUAGUUUCUAAA --------------((((((((.....-.(((.((........))))).......(((((((....)))))))..........))))))))...........-................ ( -17.70) >DroSec_CAF1 18358 114 - 1 CCCGGAAUACCACUGUCACUGGUCUAU-UGCAAGUCUUUCAUUGCUGUUUUUUUAAUAUGAAAAUC---UUAUCUAUUCUCAACCAGUGACACGCUUUAAAU-CACUGUAGUUUCUUAA ...(((((.....((((((((((....-.((((........)))).((((((.......)))))).---.............)))))))))).((.......-....)).))))).... ( -20.20) >DroSim_CAF1 18754 117 - 1 CCCGGAAUACCACUGUCACUGGUCUAU-UGCAAGUCUUUCAUUGCUGUUUUUCUAAUAUGAAAAUCUUCUUAUCUAUUCUCAACCAGUGACACGCUUUAAAU-CACUGUAGUUUCUUAA ...(((((.....((((((((((....-.((((........))))...(((((......)))))..................)))))))))).((.......-....)).))))).... ( -22.10) >DroEre_CAF1 18602 117 - 1 CCCAGAAUACCACUGACACUGGUAUAU-UGCUAGUUUUUCAGUACGGUUUUUCUAAUAGGAAAAUCUUCUUAUCUAUUCUCAACCAUUGACACA-UUUGUAUCCCCAGUAGUUACUUAA ...((((((..(((((.((((((....-.))))))...)))))..((((((((.....))))))))........)))))).(((.((((.....-..........)))).)))...... ( -20.36) >DroYak_CAF1 18825 117 - 1 CCCGGAAUACCACUGUCACUGGUCUAUUUGCAAGUUUUUCAGUACCGCUUUUCUAAUAGGAAAAUCAUCUUAUCUAU-AUCAACCAGUGACACACUUUAUGUCUCCAGUAGUU-UUUAA ...(((.......((((((((((......((..((........)).))((((((....)))))).............-....)))))))))).((.....)).))).......-..... ( -20.10) >consensus CCCGGAAUACCACUGUCACUGGUCUAU_UGCAAGUCUUUCAUUGCUGUUUUUCUAAUAGGAAAAUCUUCUUAUCUAUUCUCAACCAGUGACACGCUUUAAAU_CACUGUAGUUUCUUAA ..............(((((((((......(((.((........)))))(((((......)))))..................)))))))))............................ (-14.12 = -14.12 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:55 2006