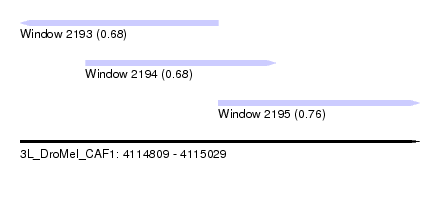
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,114,809 – 4,115,029 |
| Length | 220 |
| Max. P | 0.757058 |

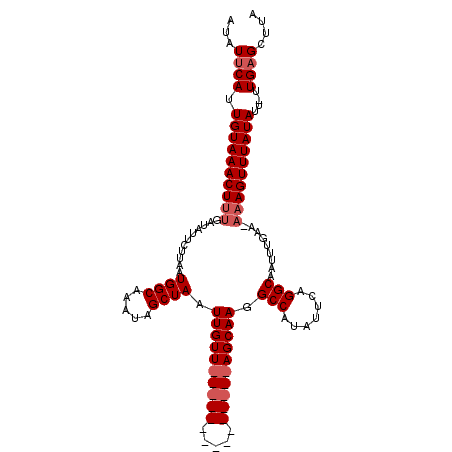
| Location | 4,114,809 – 4,114,918 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -13.77 |
| Energy contribution | -16.27 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 0.94 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4114809 109 - 23771897 GUUUUCAUUGUAAACUUUGAUAUUCUUAAUGGCAAAUAGCUAAUUGUUUUAUCGUUAUGAAGUAGCAAGGCCAUUUUGAGGCAAUUCGA----AGUUUAUAUUUUUGCGCUUA ((......((((((((((((..(((..((((((.....((((....((((((....))))))))))...))))))..))).....))))----)))))))).....))..... ( -26.40) >DroSec_CAF1 16271 96 - 1 AUAUUCAUUGUAAACUUUGAUAUUCUUAAUGGCAAAUAGCUAAUUGUU---------------AGCAA-GCCAUAUUCAGGCAAUUUGAAGAAAGUUUAUAUU-UUGAGCUUA ...((((.((((((((((..........(((((...((((.....)))---------------)....-))))).(((((.....))))).))))))))))..-.)))).... ( -20.20) >DroSim_CAF1 16662 97 - 1 AUAUUCAUUGUAAACUUUGAUAUUCUUAAUGGCAAAUAGCUAAUUGUU---------------AGCAAGGCCAUAUUCAGGCAAUUUAAAAAAAGUUUAUAUU-UUGAGCUUA ...((((.((((((((((......(((..((((((........)))))---------------)..)))(((.......))).........))))))))))..-.)))).... ( -18.80) >consensus AUAUUCAUUGUAAACUUUGAUAUUCUUAAUGGCAAAUAGCUAAUUGUU_______________AGCAAGGCCAUAUUCAGGCAAUUUGAA_AAAGUUUAUAUU_UUGAGCUUA ...((((.((((((((((...........((((.....)))).((((((((((.....)))))))))).(((.......))).........))))))))))....)))).... (-13.77 = -16.27 + 2.50)


| Location | 4,114,845 – 4,114,950 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.08 |
| Mean single sequence MFE | -20.02 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4114845 105 + 23771897 UGGCCUUGCUACUUCAUAACGAUAAAACAAUUAGCUAUUUGCCAUUAAGAAUAUCAAAGUUUACAAUGAAAACCUGCUUAAUCAUUGUGAAAUGGGUUU-UUU----CAA ((((...((((....................)))).....))))....(((.(((....((((((((((............))))))))))...)))..-.))----).. ( -17.85) >DroSec_CAF1 16310 94 + 1 UGGC-UUGCU---------------AACAAUUAGCUAUUUGCCAUUAAGAAUAUCAAAGUUUACAAUGAAUAUCUGCUUACUCAUUGUGAAAUGGUUUUUUUUUUCACAA ((((-..(((---------------(.....)))).....))))..(((((.((((...((((((((((.((......)).)))))))))).)))).)))))........ ( -21.40) >DroSim_CAF1 16701 94 + 1 UGGCCUUGCU---------------AACAAUUAGCUAUUUGCCAUUAAGAAUAUCAAAGUUUACAAUGAAUAUCUGCUUACUCAUUGUGAAAUGGGUUU-UUUUUCACAA ((((...(((---------------(.....)))).....))))..(((((.(((....((((((((((.((......)).))))))))))...))).)-))))...... ( -20.80) >consensus UGGCCUUGCU_______________AACAAUUAGCUAUUUGCCAUUAAGAAUAUCAAAGUUUACAAUGAAUAUCUGCUUACUCAUUGUGAAAUGGGUUU_UUUUUCACAA ((((...(((......................))).....)))).........(((...((((((((((.((......)).)))))))))).)))............... (-13.98 = -14.32 + 0.33)


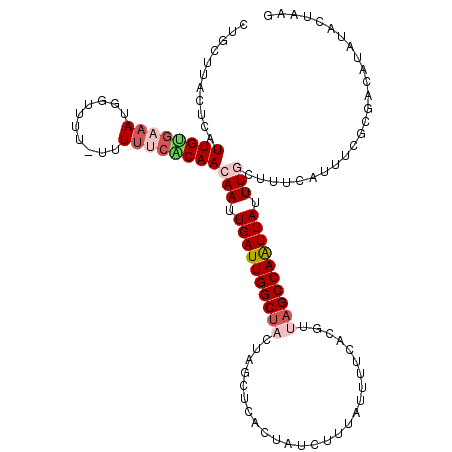
| Location | 4,114,918 – 4,115,029 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -21.91 |
| Consensus MFE | -11.93 |
| Energy contribution | -13.49 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4114918 111 + 23771897 CUGCUUAAUCAUUGUGAAAUGGGUUU-UUU----CAACAAUUGAUUGGCUACUAGCUCACUAUCUUUAGUUUCAUGUUAGCCAAUUAUUUGCUUUCAUUUCGCGACAUUUACUAAG ...........((((((((((((...-...----...(((.((((((((((.((....((((....))))....)).)))))))))).)))..))))))))))))........... ( -26.80) >DroSec_CAF1 16367 116 + 1 CUGCUUACUCAUUGUGAAAUGGUUUUUUUUUUCACAACAAUUGAUUGGCUACUAGCUCACUAUCUUUAUUUUUACGUUAGCCAAUUAUUUGCUUUCAUUUCGCGACAUCUACUAAG ...........(((((((((((...............(((.((((((((((..........................)))))))))).)))...)))))))))))........... ( -20.64) >DroSim_CAF1 16759 115 + 1 CUGCUUACUCAUUGUGAAAUGGGUUU-UUUUUCACAACAAUUGAUUGGCUACUAGCUCACUAUCUUUAUUUUUACGUUAGCCAAUUAUUUGCUUUCAUUUCGCGACAUCUACUAAG ...........(((((((((((((..-......))..(((.((((((((((..........................)))))))))).)))...)))))))))))........... ( -21.97) >DroEre_CAF1 16587 111 + 1 C---UUGUGAAUUGCGAAACUGUUU--UCUUUCGCAAAAAUUGAUUGGCGUCUAAUUCACUAUAUUUAUUUUCACAUUAGCCAAUUAUUUGCUUUUAUUUCGAUUCAUAUACUAAG .---.(((((((((.((((......--..))))(((((...((((((((..............................)))))))))))))........)))))))))....... ( -20.81) >DroYak_CAF1 16819 104 + 1 C---UUGUGAAUUGUGCAAUUGUUU--UCUUUCGCAAAAAUUGAUUGGCUCCUAGUUAACGAUAUUUACU------UUAGCCAGUUAUUUGCUUUCAUUUCGGGU-GUAUACUAAG .---........((((((.(((...--......(((((...(((((((((...(((...........)))------..))))))))))))))........))).)-)))))..... ( -19.33) >consensus CUGCUUACUCAUUGUGAAAUGGUUUU_UUUUUCACAACAAUUGAUUGGCUACUAGCUCACUAUCUUUAUUUUCACGUUAGCCAAUUAUUUGCUUUCAUUUCGCGACAUAUACUAAG ...........((((((((..........))))))))(((.((((((((((..........................)))))))))).)))......................... (-11.93 = -13.49 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:51 2006