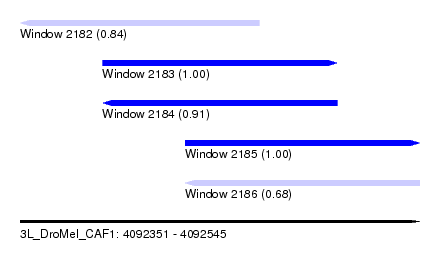
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,092,351 – 4,092,545 |
| Length | 194 |
| Max. P | 0.999122 |

| Location | 4,092,351 – 4,092,467 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -33.90 |
| Energy contribution | -34.50 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4092351 116 - 23771897 ----CGACGUCAACGCGGACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGUAAGCGAACAGGUUGUCCAGACAAAAGAUGUUGCGGCAGAUGGAAG ----.((((((......))))))((((...)))).....(((((...........)))))...(((((.((((((..((.((((..((((....))))....)))))).))))))))))) ( -36.60) >DroSec_CAF1 36417 116 - 1 ----CGACGUCAACGCGAACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCGAACAGGUUGGCCAGACAAAAGAUGUUGCGGCAGAUGGAAG ----...((((..(((.((((((((((...))))..((((((((.((((.(........)...(((((.....)))))....))))))))))))......)))))))))...)))).... ( -37.00) >DroSim_CAF1 36739 116 - 1 ----CGACGUCAACGCGGACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCGAACAGGUUGGCCAGACAAAAGAUGUUGCGGCAGAUGGAAG ----(((((((......)))))))..((((((((...(((((((.((((.(........)...(((((.....)))))....)))))))))))((((.....))))))))).)))..... ( -38.60) >DroEre_CAF1 37180 120 - 1 CCUUCGACUUCGACGCGGACGUCGCAGUCGCUGCACUUGGCGCAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCGAACAGGUUGGCCAGACAAAAGAUGUUGCGGCAGAUGGAAG ((.((((((.(((((....))))).))))(((((...((((.(((((((.(........)...(((((.....)))))....)))))))))))((((.....))))))))).)).))... ( -45.10) >DroYak_CAF1 37874 116 - 1 ----CGACGUCGACGCGGACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCGAACAGGUUGGCCAGACAAAAGAUGUUGCGGCAGAUGGAAG ----(((((((......)))))))..((((((((...(((((((.((((.(........)...(((((.....)))))....)))))))))))((((.....))))))))).)))..... ( -38.80) >consensus ____CGACGUCAACGCGGACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCGAACAGGUUGGCCAGACAAAAGAUGUUGCGGCAGAUGGAAG ....(((((((......)))))))..(((((.(((((..(......)..))....(((((......(((((.((....)).)))))...)))))........))).)))))......... (-33.90 = -34.50 + 0.60)

| Location | 4,092,391 – 4,092,505 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -47.82 |
| Consensus MFE | -40.18 |
| Energy contribution | -40.82 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4092391 114 + 23771897 CGCUUACAGACAGGAAGUUUGCCAAUUUUCUCAAGUUUAGCCAAGUGCAGCGACUGCGACGUCCGCGUUGACGUCG------ACGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCUGU .(((...((((((((((((....))))))))...))))......(((((((..((((((((((......))))))(------(((((........))))))))))..))))))))))... ( -46.60) >DroSec_CAF1 36457 114 + 1 CGCUUCCAGACAGGAAGUUUGCCAAUUUUCUCAAGUUUAGCCAAGUGCAGCGACUGCGACGUUCGCGUUGACGUCG------ACGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCAGU .((((((.....))))))((((.(((((....))))).......(((((((..((((((((((.((((((....))------)))).........))))))))))..))))))).)))). ( -46.30) >DroSim_CAF1 36779 114 + 1 CGCUUCCAGACAGGAAGUUUGCCAAUUUUCUCAAGUUUAGCCAAGUGCAGCGACUGCGACGUCCGCGUUGACGUCG------GCGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCUGU .((((((.....))))))...................((((...(((((((..((((((((((......))))))(------(((((........))))))))))..))))))).)))). ( -48.80) >DroEre_CAF1 37220 120 + 1 CGCUUCCAGACAGGAAGUUUGCCAAUUUUCUCAAGUUGCGCCAAGUGCAGCGACUGCGACGUCCGCGUCGAAGUCGAAGGCGAAGCUCCUCUGCUAGCGUCGCAGCAGCUGCGCAGCUGU .((((((.....))))))..((((((((....)))))).))...(((((((..(((((((((.(((.(((....)))..))).(((......))).)))))))))..)))))))...... ( -52.60) >DroYak_CAF1 37914 114 + 1 CGCUUCCAGACAGGAAGUUUGCCAAUUUUCUCAAGUUUAGCCAAGUGCAGCGACUGCGACGUCCGCGUCGACGUCG------AAGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCUGU .((((((.....))))))...................((((...(((((((..(((((((((..((.(((....))------).........))..)))))))))..))))))).)))). ( -44.80) >consensus CGCUUCCAGACAGGAAGUUUGCCAAUUUUCUCAAGUUUAGCCAAGUGCAGCGACUGCGACGUCCGCGUUGACGUCG______ACGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCUGU .((((((.....)))))).....................((...(((((((..(((((((((..(((..(((((........))).))..).))..)))))))))..))))))).))... (-40.18 = -40.82 + 0.64)

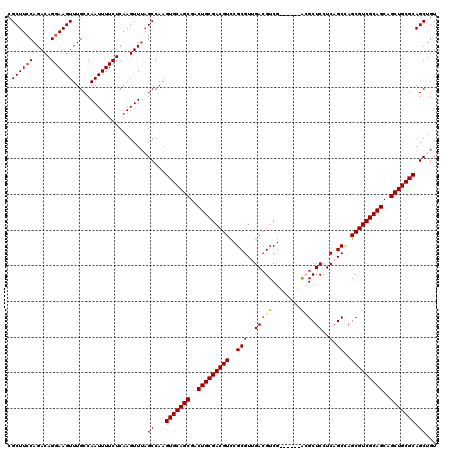
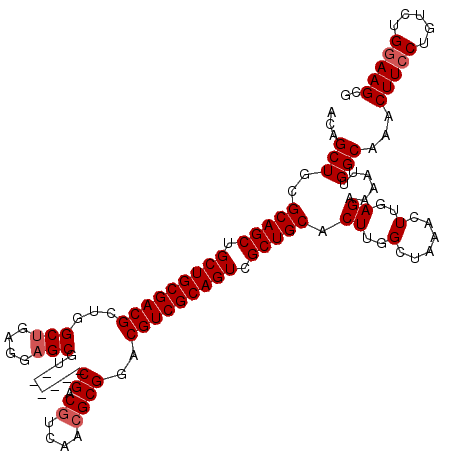
| Location | 4,092,391 – 4,092,505 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -45.82 |
| Consensus MFE | -39.22 |
| Energy contribution | -39.82 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4092391 114 - 23771897 ACAGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCGU------CGACGUCAACGCGGACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGUAAGCG ...((((((((((.(((((((((((........)))))------)((((((......))))))))))).))))).((..(......)..))......((((.......)))).)).))). ( -45.70) >DroSec_CAF1 36457 114 - 1 ACUGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCGU------CGACGUCAACGCGAACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCG ..((((..(((((.(((((((((((........)))))------)(((((........)))))))))).))))).((..(......)..))......))))..(((((.....))))).. ( -46.00) >DroSim_CAF1 36779 114 - 1 ACAGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCGC------CGACGUCAACGCGGACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCG ...(((..(((((.(((((((((...(((....))).(------((.((....))))).))))))))).))))).((..(......)..))......)))...(((((.....))))).. ( -45.90) >DroEre_CAF1 37220 120 - 1 ACAGCUGCGCAGCUGCUGCGACGCUAGCAGAGGAGCUUCGCCUUCGACUUCGACGCGGACGUCGCAGUCGCUGCACUUGGCGCAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCG ...(((..(((((.(((((((((...((......))(((((..(((....))).)))))))))))))).))))).((..(......)..))......)))...(((((.....))))).. ( -46.20) >DroYak_CAF1 37914 114 - 1 ACAGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCUU------CGACGUCGACGCGGACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCG ...(((..(((((.(((((((((((((((....))).(------((....)))..))).))))))))).))))).((..(......)..))......)))...(((((.....))))).. ( -45.30) >consensus ACAGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCGU______CGACGUCAACGCGGACGUCGCAGUCGCUGCACUUGGCUAAACUUGAGAAAAUUGGCAAACUUCCUGUCUGGAAGCG ...(((..(((((.(((((((((...(((....)))........((.((....))))..))))))))).))))).((..(......)..))......)))...(((((.....))))).. (-39.22 = -39.82 + 0.60)

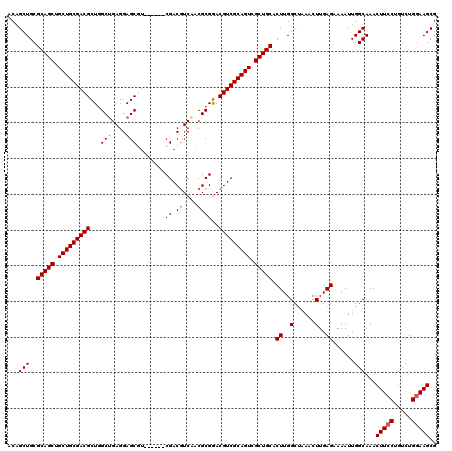
| Location | 4,092,431 – 4,092,545 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -44.08 |
| Consensus MFE | -37.46 |
| Energy contribution | -38.10 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4092431 114 + 23771897 CCAAGUGCAGCGACUGCGACGUCCGCGUUGACGUCG------ACGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCUGUUUAAUAUUCGGUGAGUAUUUCGAAAAACAAAGAAAAACCC ....(((((((..((((((((((......))))))(------(((((........))))))))))..)))))))...(((((....(((((........))))))))))........... ( -44.80) >DroSec_CAF1 36497 114 + 1 CCAAGUGCAGCGACUGCGACGUUCGCGUUGACGUCG------ACGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCAGUUUAAUAUUCGGUGAGUAUUUCCAAAAACAAAGGAAAACCC ((..(((((((..((((((((((.((((((....))------)))).........))))))))))..)))))))....(((((((((((...))))))).....))))...))....... ( -42.90) >DroSim_CAF1 36819 114 + 1 CCAAGUGCAGCGACUGCGACGUCCGCGUUGACGUCG------GCGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCUGUUUAAUAUUCGGUGAGUAUUUCCAAAAACAAAGGAAAACCC ((..(((((((..((((((((((......))))))(------(((((........))))))))))..)))))))...((((((((((((...))))))).....)))))..))....... ( -44.80) >DroEre_CAF1 37260 120 + 1 CCAAGUGCAGCGACUGCGACGUCCGCGUCGAAGUCGAAGGCGAAGCUCCUCUGCUAGCGUCGCAGCAGCUGCGCAGCUGUUUAAUAUUCGGUGAGUAUUUCCAAAAACAAAGGAAAACCC ((..(((((((..(((((((((.(((.(((....)))..))).(((......))).)))))))))..)))))))...((((((((((((...))))))).....)))))..))....... ( -46.70) >DroYak_CAF1 37954 114 + 1 CCAAGUGCAGCGACUGCGACGUCCGCGUCGACGUCG------AAGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCUGUUUAAUAUUCGGUGAGUAUUUCCAAAAACAAAGGAAAGCCC ....(((((((..(((((((((..((.(((....))------).........))..)))))))))..))))))).(((.....((((((...))))))((((.........))))))).. ( -41.20) >consensus CCAAGUGCAGCGACUGCGACGUCCGCGUUGACGUCG______ACGCUCCUCAGCCAGCGUCGCAGCAGCUGCGCAGCUGUUUAAUAUUCGGUGAGUAUUUCCAAAAACAAAGGAAAACCC ((..(((((((..(((((((((..(((..(((((........))).))..).))..)))))))))..)))))))....(((((((((((...))))))).....))))...))....... (-37.46 = -38.10 + 0.64)

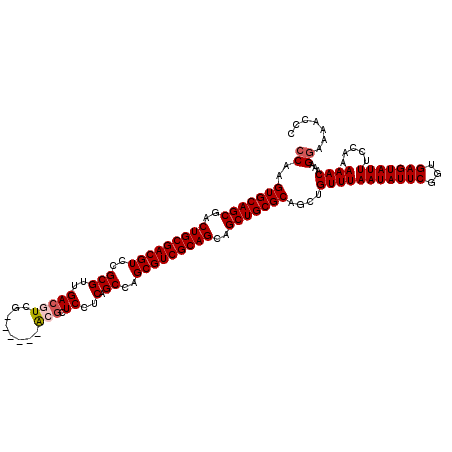
| Location | 4,092,431 – 4,092,545 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -42.56 |
| Consensus MFE | -35.56 |
| Energy contribution | -36.20 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4092431 114 - 23771897 GGGUUUUUCUUUGUUUUUCGAAAUACUCACCGAAUAUUAAACAGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCGU------CGACGUCAACGCGGACGUCGCAGUCGCUGCACUUGG ((((.((((..........)))).))))....................(((((.(((((((((((........)))))------)((((((......))))))))))).)))))...... ( -43.70) >DroSec_CAF1 36497 114 - 1 GGGUUUUCCUUUGUUUUUGGAAAUACUCACCGAAUAUUAAACUGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCGU------CGACGUCAACGCGAACGUCGCAGUCGCUGCACUUGG (((((((((.........))))).))))....................(((((.(((((((((((........)))))------)(((((........)))))))))).)))))...... ( -42.50) >DroSim_CAF1 36819 114 - 1 GGGUUUUCCUUUGUUUUUGGAAAUACUCACCGAAUAUUAAACAGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCGC------CGACGUCAACGCGGACGUCGCAGUCGCUGCACUUGG (((((((((.........))))).))))....................(((((.(((((((((...(((....))).(------((.((....))))).))))))))).)))))...... ( -43.00) >DroEre_CAF1 37260 120 - 1 GGGUUUUCCUUUGUUUUUGGAAAUACUCACCGAAUAUUAAACAGCUGCGCAGCUGCUGCGACGCUAGCAGAGGAGCUUCGCCUUCGACUUCGACGCGGACGUCGCAGUCGCUGCACUUGG (((((((((.........))))).))))....................(((((.(((((((((...((......))(((((..(((....))).)))))))))))))).)))))...... ( -43.30) >DroYak_CAF1 37954 114 - 1 GGGCUUUCCUUUGUUUUUGGAAAUACUCACCGAAUAUUAAACAGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCUU------CGACGUCGACGCGGACGUCGCAGUCGCUGCACUUGG .((((....((((..(((((.........)))))...)))).))))..(((((.(((((((((((((((....))).(------((....)))..))).))))))))).)))))...... ( -40.30) >consensus GGGUUUUCCUUUGUUUUUGGAAAUACUCACCGAAUAUUAAACAGCUGCGCAGCUGCUGCGACGCUGGCUGAGGAGCGU______CGACGUCAACGCGGACGUCGCAGUCGCUGCACUUGG (((((((((.........))))).))))....................(((((.(((((((((...(((....)))........((.((....))))..))))))))).)))))...... (-35.56 = -36.20 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:42 2006