
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,070,175 – 4,070,308 |
| Length | 133 |
| Max. P | 0.832564 |

| Location | 4,070,175 – 4,070,286 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.39 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -26.61 |
| Energy contribution | -26.92 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4070175 111 + 23771897 AGAGUUCCACCCACUCCACUUCCAUUCUUAUCUGACACACGGAGCAGCUGGCGUAGGAUGUUACCACACACUUGGCACUUGGCACUUGGUCCUGCUGCUGCUCCAGCUCCU .(((((...................((......)).....((((((((....(((((((((....)).((..((.(....).))..))))))))).))))))))))))).. ( -30.50) >DroSec_CAF1 14364 111 + 1 AGAGUUCCACCCACACCACUCCCAUUCUUAUCUGACACACGGAGCAGCUGGCGGAGGAUGUUACCACACACUUGGCACUUGGCACUUGGUCCUGCUGCUGCUCCAGCUCCU .(((((...................((......)).....((((((((..((.(((..(((....)))..))).))....((((........))))))))))))))))).. ( -31.50) >DroSim_CAF1 14501 111 + 1 AGAGUUCCACCCACACCACUCCCAUUCUUAUCUGACACACGGAGCAGCUGGCGGAGGAUGUUACCACACACUUGGCACUUGGCACUUGGUCCUGCUGCUGCUCCAGCUCCU .(((((...................((......)).....((((((((..((.(((..(((....)))..))).))....((((........))))))))))))))))).. ( -31.50) >DroEre_CAF1 15270 91 + 1 AGAGUUUCUCCCAC-----------ACUUAUCUGACACACGGAGCAGCUGGCGAAGGAUGUUACCACA---------CUUGGCACUUGGUCCUGCCGCUGCUCCUGCUCCU .((((........(-----------(......))......((((((((.(((..(((((....(((..---------..)))......)))))))))))))))).)))).. ( -29.00) >consensus AGAGUUCCACCCACACCACUCCCAUUCUUAUCUGACACACGGAGCAGCUGGCGGAGGAUGUUACCACACACUUGGCACUUGGCACUUGGUCCUGCUGCUGCUCCAGCUCCU .(((((...................((......)).....((((((((.(((..(((((....(((.............)))......))))))))))))))))))))).. (-26.61 = -26.92 + 0.31)

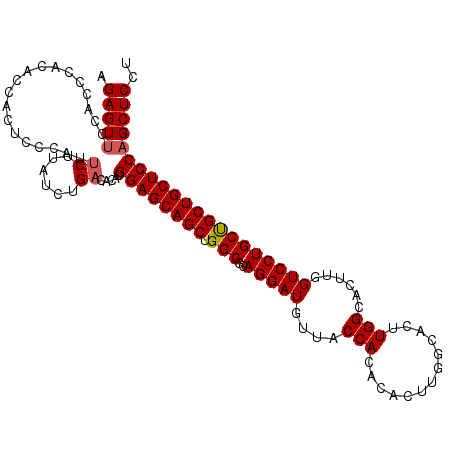

| Location | 4,070,212 – 4,070,308 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -31.33 |
| Energy contribution | -31.14 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4070212 96 + 23771897 CACGGAGCAGCUGGCGUAGGAUGUUACCACACACUUGGCACUUGGCACUUGGUCCUGCUGCUGCUCCAGCUCCU--GCUCCCUGGGCAGGAUCCAAUG ...((((((((....(((((((((....)).((..((.(....).))..))))))))).))))))))...((((--(((.....)))))))....... ( -39.10) >DroSec_CAF1 14401 96 + 1 CACGGAGCAGCUGGCGGAGGAUGUUACCACACACUUGGCACUUGGCACUUGGUCCUGCUGCUGCUCCAGCUCCU--GCUCCUUGGGCAGGAUCCACUG ...((((((((..((.(((..(((....)))..))).))....((((........))))))))))))...((((--(((.....)))))))....... ( -40.10) >DroSim_CAF1 14538 96 + 1 CACGGAGCAGCUGGCGGAGGAUGUUACCACACACUUGGCACUUGGCACUUGGUCCUGCUGCUGCUCCAGCUCCU--GCUCCUGGGGCAGGAUCCACUG ...((((((((..((.(((..(((....)))..))).))....((((........))))))))))))...((((--(((.....)))))))....... ( -40.10) >DroEre_CAF1 15296 89 + 1 CACGGAGCAGCUGGCGAAGGAUGUUACCACA---------CUUGGCACUUGGUCCUGCCGCUGCUCCUGCUCCUUGGCUCCUUGGGCAGGAUCCACUG ...((((((((.(((..(((((....(((..---------..)))......)))))))))))))((((((.((..(....)..))))))))))).... ( -37.10) >consensus CACGGAGCAGCUGGCGGAGGAUGUUACCACACACUUGGCACUUGGCACUUGGUCCUGCUGCUGCUCCAGCUCCU__GCUCCUUGGGCAGGAUCCACUG ...((((((((.(((..(((((....(((.............)))......))))))))))))))))...((((..(((.....)))))))....... (-31.33 = -31.14 + -0.19)

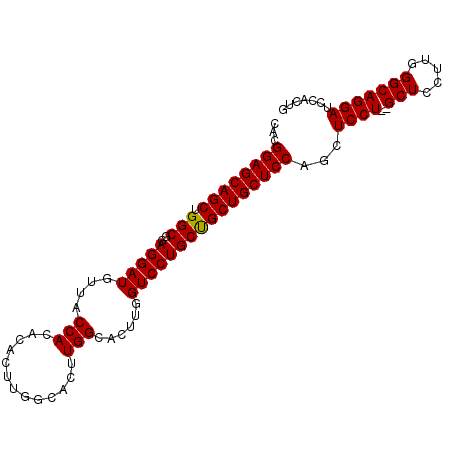
| Location | 4,070,212 – 4,070,308 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -39.33 |
| Consensus MFE | -30.66 |
| Energy contribution | -30.73 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4070212 96 - 23771897 CAUUGGAUCCUGCCCAGGGAGC--AGGAGCUGGAGCAGCAGCAGGACCAAGUGCCAAGUGCCAAGUGUGUGGUAACAUCCUACGCCAGCUGCUCCGUG ....((.(((((((....).))--)))).))((((((((.((((((.....(((((.(..(...)..).)))))...))))..))..))))))))... ( -39.20) >DroSec_CAF1 14401 96 - 1 CAGUGGAUCCUGCCCAAGGAGC--AGGAGCUGGAGCAGCAGCAGGACCAAGUGCCAAGUGCCAAGUGUGUGGUAACAUCCUCCGCCAGCUGCUCCGUG ....((.(((((((....).))--)))).))((((((((.((((((.....(((((.(..(...)..).)))))...))))..))..))))))))... ( -39.20) >DroSim_CAF1 14538 96 - 1 CAGUGGAUCCUGCCCCAGGAGC--AGGAGCUGGAGCAGCAGCAGGACCAAGUGCCAAGUGCCAAGUGUGUGGUAACAUCCUCCGCCAGCUGCUCCGUG ....((.(((((((....).))--)))).))((((((((.((((((.....(((((.(..(...)..).)))))...))))..))..))))))))... ( -39.20) >DroEre_CAF1 15296 89 - 1 CAGUGGAUCCUGCCCAAGGAGCCAAGGAGCAGGAGCAGCGGCAGGACCAAGUGCCAAG---------UGUGGUAACAUCCUUCGCCAGCUGCUCCGUG (..(((.((((.....)))).)))..)....(((((((((((((((.....(((((..---------..)))))...))))..))).))))))))... ( -39.70) >consensus CAGUGGAUCCUGCCCAAGGAGC__AGGAGCUGGAGCAGCAGCAGGACCAAGUGCCAAGUGCCAAGUGUGUGGUAACAUCCUCCGCCAGCUGCUCCGUG .......(((((((....).))..))))...((((((((.((((((.....(((((.(..(...)..).)))))...))))..))..))))))))... (-30.66 = -30.73 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:31 2006