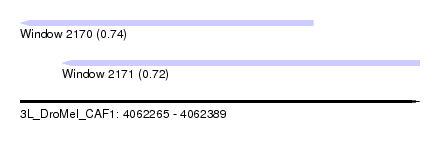
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,062,265 – 4,062,389 |
| Length | 124 |
| Max. P | 0.740520 |

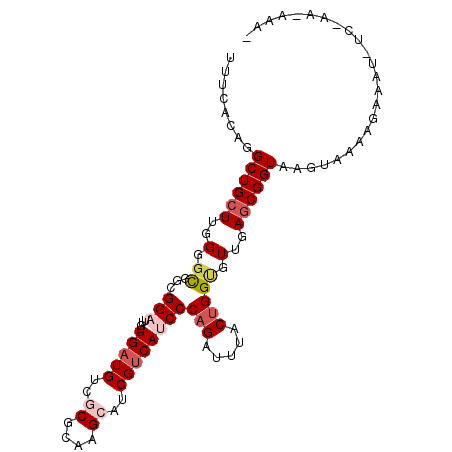
| Location | 4,062,265 – 4,062,356 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -21.35 |
| Energy contribution | -22.38 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4062265 91 - 23771897 UUUC-CAGGCUGCUUGGGGCGGUGGAUUGUUGAUGUCGCGUAAGCAUCGUCAUCCCAGAUUUACUGGUGUUGAGCGGUA--------AGUU-UUUAGAAAA- ....-...((((((..(.((((((((((..(((((..((....))..))))).....)))))))).)).)..)))))).--------....-.........- ( -31.00) >DroVir_CAF1 8912 97 - 1 UCUAACAGGCUGCUUGGGGCGGUGGAUUGCUGAUGUCGCGCAAGCAUCGCCAUCCCAGAUUCACAGGCGUUGAACGGUAAGUACUAUCUAG-UC----UAAU ......((((((((((.(((((((..((((((....)).)))).)))))))(((...)))...))))).......((((.....)))).))-))----)... ( -26.70) >DroPse_CAF1 6492 100 - 1 UUUUAAAGGCUGCUUGGGGCGGCGGUCUGUUGAUGUCUCGCAAGCAUCGUCAUCCCAGAUUCACUGGCGUUGAGCGGUAAGUAGAAGAAAU-UC-AAGAGGU ........((((((..(.((((.((((((.(((((...(....)...)))))...)))).)).)).)).)..)))))).....(((....)-))-....... ( -30.80) >DroEre_CAF1 6580 101 - 1 UUUCACAGGCUGCUUGGGGCGGCGGAUUGUUGAUGUCGCGUAAGCAUCGUCAUCCCAGAUUUACUGGUGUUGAGCGGUAAGUAAGAGAGUUUUUUAAGAAA- ((((((..((((((..(.((((..((((..(((((..((....))..))))).....))))..)).)).)..))))))..))..)))).............- ( -30.10) >DroWil_CAF1 6668 100 - 1 UCCUACAGGCUGCUUGGGGUGGUGGCCUAAUGAUGUCCCGCAAGCAUCGUCAUCCCAGAUUUACUGGAGUUGAGCGGUAAGUUCAAUGAAU-AC-UAUAAGU ......((((..((......))..)))).((((((...(....)...)))))).((((.....)))).(((((((.....)))))))....-..-....... ( -29.20) >DroYak_CAF1 6669 87 - 1 UUUCACAGGCUGCUUGGGGCGGCGGAUUGUUGAUGUCGCGUAAGCAUCGUCAUCCCAGAUUUACUGGUGUUGAGCGGUAAGUAAGAG--------------- ....((..((((((..(.((((..((((..(((((..((....))..))))).....))))..)).)).)..))))))..)).....--------------- ( -29.30) >consensus UUUCACAGGCUGCUUGGGGCGGCGGAUUGUUGAUGUCGCGCAAGCAUCGUCAUCCCAGAUUUACUGGUGUUGAGCGGUAAGUAAAAGAAAU_UC_AA_AAA_ ........((((((..(.((...(((....(((((..((....))..))))))))(((.....))))).)..))))))........................ (-21.35 = -22.38 + 1.03)


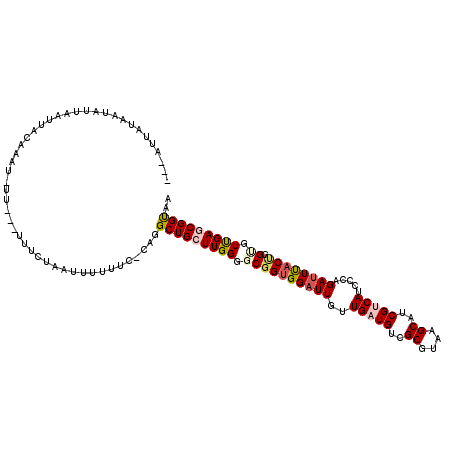
| Location | 4,062,278 – 4,062,389 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -24.96 |
| Energy contribution | -25.77 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4062278 111 - 23771897 ---AUUGUAAUAUUAAUUACUAAU-UU---UUUCUGAUUUUUUC-CAGGCUGCUUGGGGCGGUGGAUUGUUGAUGUCGCGUAAGCAUCGUCAUCCCAGAUUUACUGGUGUUGAGCGGUA- ---...(((((....)))))....-..---...(((........-)))((((((..(.((((((((((..(((((..((....))..))))).....)))))))).)).)..)))))).- ( -34.10) >DroVir_CAF1 8929 111 - 1 ---UUGA--AUAUUUAAUAGAGUU-UUCUGUUCG---UGCUCUAACAGGCUGCUUGGGGCGGUGGAUUGCUGAUGUCGCGCAAGCAUCGCCAUCCCAGAUUCACAGGCGUUGAACGGUAA ---....--........((((((.-.........---.))))))....((((.(..(.((.(((((((...((((.(((....))...).))))...)))))))..)).)..).)))).. ( -30.10) >DroGri_CAF1 6693 115 - 1 ---CUUAACUUUUCUAAUGCAAUU-UUUGGUUUCAA-UUUCUUUACAGGCUGCUUGGGGCGGUGGUUUGUUGAUGUCGCGAAAGCAUCGCCAUCCCAGAUUCACCGGAAUCGAACGGCAA ---..............(((....-(((((((((..-...........(((......)))(((((((((..((((..((((.....)))))))).)))).))))))))))))))..))). ( -29.40) >DroSec_CAF1 6642 112 - 1 ---AUUGUAAUAUUAAUGACCAAU-UU---UUUCUUAUUUUUUC-CAGGCUGCUUGGGGCGGUGGAUUGUUGAUGUCGCGUAAGCAUCGUCAUCCCAGAUUUACUGGGGUUGAGCGGUAA ---...............(((...-..---............((-(..(((((.....))))))))..(((((((..((....))..))))(((((((.....)))))))..)))))).. ( -30.60) >DroSim_CAF1 6636 112 - 1 ---AUUAUAAUAUUAAUGACAAAU-UU---UUUGUAAUUUUUUC-UAGGCUGCUUGGGGCGGUGGAUUGUUGAUGUCGCGUAAGCAUCGUCAUCCCAGAUUUACUGGUGUUGAGCGGUGA ---(((((((....(((.....))-).---.)))))))......-...((((((..(.((((((((((..(((((..((....))..))))).....)))))))).)).)..)))))).. ( -32.00) >DroEre_CAF1 6601 116 - 1 CAUAUUU-UAUAUUCAUUCCAAAUUUU---UUUAAUUUUUUUUCACAGGCUGCUUGGGGCGGCGGAUUGUUGAUGUCGCGUAAGCAUCGUCAUCCCAGAUUUACUGGUGUUGAGCGGUAA .......-...................---..................((((((..(.((((..((((..(((((..((....))..))))).....))))..)).)).)..)))))).. ( -28.20) >consensus ___AUUAUAAUAUUAAUUACAAAU_UU___UUUCUAAUUUUUUC_CAGGCUGCUUGGGGCGGUGGAUUGUUGAUGUCGCGUAAGCAUCGUCAUCCCAGAUUUACUGGUGUUGAGCGGUAA ................................................(((((((((.((((((((((..(((((..((....))..))))).....)))))))).)).))))))))).. (-24.96 = -25.77 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:28 2006