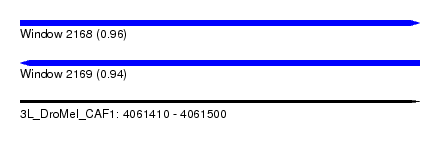
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,061,410 – 4,061,500 |
| Length | 90 |
| Max. P | 0.957313 |

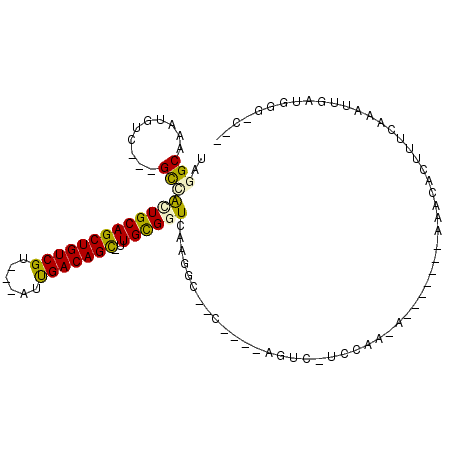
| Location | 4,061,410 – 4,061,500 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.89 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -11.37 |
| Energy contribution | -12.15 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4061410 90 + 23771897 AUG-GCAAUCAAUUUGAAAGUGUUC-------U-GUGGA-GUCG----GC-UGCUUGACCGCAA-GCUGUCGAUUCCACGACAGCUGCACUUGC---CGCAUUUGCCUA ..(-((((............((((.-------.-(((((-((((----((-.(((((....)))-)).)))))))))))))))((.((....))---.))..))))).. ( -38.50) >DroVir_CAF1 5636 82 + 1 --G-C-AAUCAAUUUGAAAGUGUUU-------A-UGGGA-CACU-------GCCUUGACCGCAA-GCUGUCAAU---ACGACAGCUGCAGUGGC---GACAUUUGCCUA --(-(-((..........((((((.-------.-...))-))))-------(((...((.(((.-((((((...---..))))))))).)))))---.....))))... ( -26.70) >DroGri_CAF1 5876 85 + 1 --GGUCCAUCAAUCUGAAAGUGUUU-------A-UGGGAGCACU-------CCCUUGACCGCAA-ACUGUCAAU---ACGACAGCUGCAGUGGC---GACAUUUGCCUA --((((..((.....))........-------.-.((((....)-------)))..))))(((.-.(((((...---..))))).)))...(((---((...))))).. ( -24.30) >DroWil_CAF1 5667 97 + 1 UAG-CCAAUCAAUUUGAAAGUGUUUAUGGCCUUAAUGGA-GCU-CUAGG--GCCUUGACCACAA-GCUGUCAAU---ACGACAGCUGCACUUAC---GACAUUUGCCUA (((-.(((.....(((.(((((((((.(((((((.....-...-.))))--))).))).....(-((((((...---..))))))))))))).)---))...))).))) ( -28.30) >DroMoj_CAF1 6489 90 + 1 --G-CCCAUCAAUUUGAAAGUGUUU-------A-UGGGA-CACU----UCUCCCUUGACCGCAAAACUGUCAAU---ACGACAGCUGCAACGGCGGUGGCAUUUGCCUA --(-((..((((...(((((((((.-------.-...))-))))----).))..))))..(((...(((((...---..))))).)))...)))...(((....))).. ( -25.80) >DroAna_CAF1 5153 88 + 1 AAG-GCCAUCAAUUUGAAACUGUU--------U-AUGGA-GUCU----GG-AG-UUGACCGCAG-GCUGUCAAUACGACGACAGCUGCAAUAGC---CACAUUUGCCUA .((-((..(((((((.(.(((.(.--------.-..).)-)).)----.)-))-))))..((((-.(((((........)))))))))......---.......)))). ( -27.00) >consensus __G_CCAAUCAAUUUGAAAGUGUUU_______A_UGGGA_CACU____G__CCCUUGACCGCAA_GCUGUCAAU___ACGACAGCUGCAAUGGC___GACAUUUGCCUA .................(((((((............((....................))(((..((((((........))))))))).........)))))))..... (-11.37 = -12.15 + 0.78)

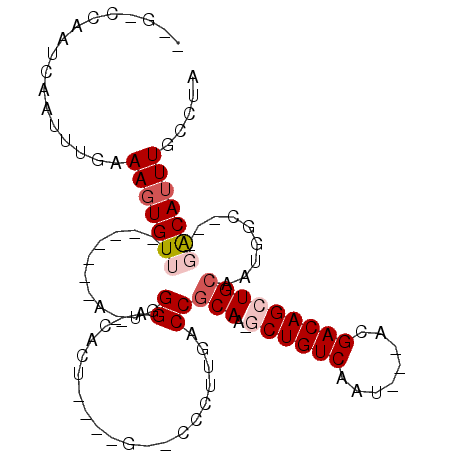
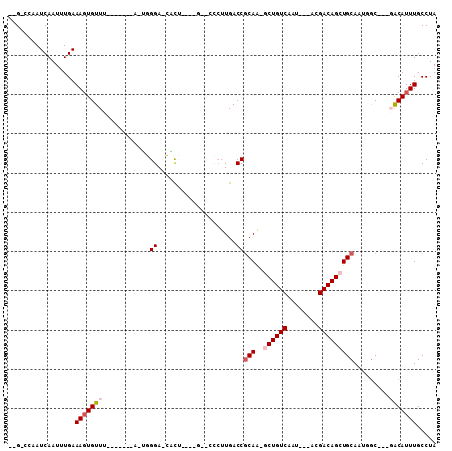
| Location | 4,061,410 – 4,061,500 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.89 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -14.65 |
| Energy contribution | -14.35 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4061410 90 - 23771897 UAGGCAAAUGCG---GCAAGUGCAGCUGUCGUGGAAUCGACAGC-UUGCGGUCAAGCA-GC----CGAC-UCCAC-A-------GAACACUUUCAAAUUGAUUGC-CAU ..(((((...((---((......((((((((......)))))))-)(((......)))-))----))..-.....-.-------(((....))).......))))-).. ( -30.60) >DroVir_CAF1 5636 82 - 1 UAGGCAAAUGUC---GCCACUGCAGCUGUCGU---AUUGACAGC-UUGCGGUCAAGGC-------AGUG-UCCCA-U-------AAACACUUUCAAAUUGAUU-G-C-- ...((((.....---(((((((((((((((..---...))))))-.))))))...)))-------((((-(....-.-------..)))))..........))-)-)-- ( -27.40) >DroGri_CAF1 5876 85 - 1 UAGGCAAAUGUC---GCCACUGCAGCUGUCGU---AUUGACAGU-UUGCGGUCAAGGG-------AGUGCUCCCA-U-------AAACACUUUCAGAUUGAUGGACC-- ..(((.......---)))((((((((((((..---...))))).-)))))))...(((-------(....)))).-.-------.......((((......))))..-- ( -26.20) >DroWil_CAF1 5667 97 - 1 UAGGCAAAUGUC---GUAAGUGCAGCUGUCGU---AUUGACAGC-UUGUGGUCAAGGC--CCUAG-AGC-UCCAUUAAGGCCAUAAACACUUUCAAAUUGAUUGG-CUA ..(((....(((---(.(((((..((((((..---...))))))-((((((((..(((--.....-.))-).......)))))))).)))))......))))..)-)). ( -25.90) >DroMoj_CAF1 6489 90 - 1 UAGGCAAAUGCCACCGCCGUUGCAGCUGUCGU---AUUGACAGUUUUGCGGUCAAGGGAGA----AGUG-UCCCA-U-------AAACACUUUCAAAUUGAUGGG-C-- ..(((....)))...(((..((((((((((..---...))))))..))))(((((..((((----.(((-(....-.-------..))))))))...))))).))-)-- ( -29.30) >DroAna_CAF1 5153 88 - 1 UAGGCAAAUGUG---GCUAUUGCAGCUGUCGUCGUAUUGACAGC-CUGCGGUCAA-CU-CC----AGAC-UCCAU-A--------AACAGUUUCAAAUUGAUGGC-CUU .((((...((((---(.....((((((((((......)))))).-))))((((..-..-..----.)))-)))))-)--------..((((.....))))...))-)). ( -27.40) >consensus UAGGCAAAUGUC___GCCACUGCAGCUGUCGU___AUUGACAGC_UUGCGGUCAAGGC__C____AGUC_UCCAA_A_______AAACACUUUCAAAUUGAUGGG_C__ ..(((..........)))(((((((((((((......)))))))..))))))......................................................... (-14.65 = -14.35 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:26 2006