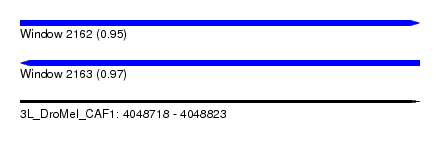
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,048,718 – 4,048,823 |
| Length | 105 |
| Max. P | 0.973347 |

| Location | 4,048,718 – 4,048,823 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.85 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -17.63 |
| Energy contribution | -17.85 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4048718 105 + 23771897 -AUCG---AUGUUCUGCUGCAUUCGAUUAGUUUAUAGCUCCAGCAGCAGAACCUCCUCAUGCUAGAGAAUGUUGAGCAGGGA---GGAGUCCAUCAUCCGAUCGUCGUACAA -..((---((((((((((((....(((((.....))).))..))))))))))((((((.((((.((.....)).))))..))---))))..............))))..... ( -32.50) >DroVir_CAF1 4847 98 + 1 -C-----ACAGUUCCGCUGCCGC-----GCUUUAUAGCUCCAGCAGCAGAACCUCCUCAUGCUAGAGUAUAUUCAGCAGCGA---UGAGUCCAUCAUGCGAUCAUCGUACAG -.-----...((((.(((((.(.-----(((....))).)..))))).)))).......((((.(((....)))))))((((---(((.(((.....).))))))))).... ( -28.60) >DroGri_CAF1 5048 97 + 1 GCUAC--ACGGUUCUGCUGCC----------UUAUAGCUCUAGCAGCAGAACUUCCUCAUGCUAGAGUAUAUUUAGCAGAGA---UGAGUCCAUCAUGCGAUCAUCAUACAA ((...--..(((((((((((.----------...........)))))))))))..(((.(((((((.....))))))))))(---(((.....))))))............. ( -30.50) >DroWil_CAF1 2943 107 + 1 -UUUC---AAGUUCUGCUGCUGUC-AUUGAUUUAGAGCUCCAACAGCAGAACCUCUUCAUGCUAGAGUAUAUUGAGUAGAGAUGAUGAGUCCAUCAUGCGAUCAUCGUAGAG -((((---..(((((((((.((..-.((......))....)).)))))))))(((...((((....))))...)))..))))(((((....)))))((((.....))))... ( -26.50) >DroMoj_CAF1 5646 108 + 1 -CUAUAUGGAGUUCUGCUGCCGAUGAUAGUCUUAAAGCUCCAGCAGCAGAACCUCCUCAUGCUAGAGUAUAUUUAACAGCGA---CGAGUCCAUCAUGCGAUCAUCGUAUAG -....(((((((((((((((...((..(((......))).))))))))))))(((.((.((.((((.....)))).))..))---.)))))))).(((((.....))))).. ( -30.10) >DroAna_CAF1 2666 94 + 1 -GACU---GUGUUCUGCUGC-----------UUAUAGCUCCAGCAGCAGAACCUCCUCAUGCUAGAGAAUAUUCAGCAGCGA---UGAGUCCAUCAUCCUGUCGUCGUACAA -(((.---..((((((((((-----------(.........)))))))))))...(((......)))........((((.((---(((.....))))))))).)))...... ( -31.70) >consensus _CUAC___AAGUUCUGCUGCCG______G_UUUAUAGCUCCAGCAGCAGAACCUCCUCAUGCUAGAGUAUAUUCAGCAGCGA___UGAGUCCAUCAUGCGAUCAUCGUACAA ..........((((((((((......................))))))))))(((.((.((((.((.....)).))))..))....)))....................... (-17.63 = -17.85 + 0.22)



| Location | 4,048,718 – 4,048,823 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.85 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -18.73 |
| Energy contribution | -19.23 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4048718 105 - 23771897 UUGUACGACGAUCGGAUGAUGGACUCC---UCCCUGCUCAACAUUCUCUAGCAUGAGGAGGUUCUGCUGCUGGAGCUAUAAACUAAUCGAAUGCAGCAGAACAU---CGAU- .....((((.(((....))).).((((---((..((((...........)))).))))))((((((((((......................)))))))))).)---))..- ( -32.45) >DroVir_CAF1 4847 98 - 1 CUGUACGAUGAUCGCAUGAUGGACUCA---UCGCUGCUGAAUAUACUCUAGCAUGAGGAGGUUCUGCUGCUGGAGCUAUAAAGC-----GCGGCAGCGGAACUGU-----G- ..(((((((((((........)).)))---))).)))........(((......)))..(((((((((((((..(((....)))-----.)))))))))))))..-----.- ( -37.80) >DroGri_CAF1 5048 97 - 1 UUGUAUGAUGAUCGCAUGAUGGACUCA---UCUCUGCUAAAUAUACUCUAGCAUGAGGAAGUUCUGCUGCUAGAGCUAUAA----------GGCAGCAGAACCGU--GUAGC .............(((((.........---(((((((((.........))))).))))..(((((((((((..........----------))))))))))))))--))... ( -30.40) >DroWil_CAF1 2943 107 - 1 CUCUACGAUGAUCGCAUGAUGGACUCAUCAUCUCUACUCAAUAUACUCUAGCAUGAAGAGGUUCUGCUGUUGGAGCUCUAAAUCAAU-GACAGCAGCAGAACUU---GAAA- ......(((((((........)).)))))........((((....((((.......))))((((((((((((...............-..))))))))))))))---))..- ( -29.13) >DroMoj_CAF1 5646 108 - 1 CUAUACGAUGAUCGCAUGAUGGACUCG---UCGCUGUUAAAUAUACUCUAGCAUGAGGAGGUUCUGCUGCUGGAGCUUUAAGACUAUCAUCGGCAGCAGAACUCCAUAUAG- .(((.((.....)).)))((((((((.---(((.(((((.........)))))))).)))((((((((((((((((.....).))....))))))))))))))))))....- ( -34.70) >DroAna_CAF1 2666 94 - 1 UUGUACGACGACAGGAUGAUGGACUCA---UCGCUGCUGAAUAUUCUCUAGCAUGAGGAGGUUCUGCUGCUGGAGCUAUAA-----------GCAGCAGAACAC---AGUC- ...........((((((((.....)))---)).)))(((....(((((......))))).(((((((((((.........)-----------)))))))))).)---))..- ( -33.30) >consensus CUGUACGAUGAUCGCAUGAUGGACUCA___UCGCUGCUAAAUAUACUCUAGCAUGAGGAGGUUCUGCUGCUGGAGCUAUAAA_C______CGGCAGCAGAACUC___AGAG_ ...........((.((((.((((.......................)))).)))).))..(((((((((((....................))))))))))).......... (-18.73 = -19.23 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:20 2006