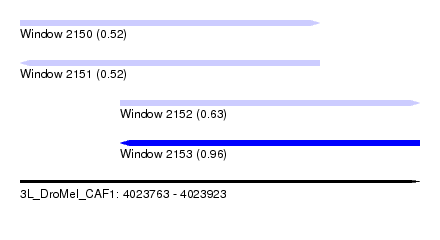
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,023,763 – 4,023,923 |
| Length | 160 |
| Max. P | 0.955275 |

| Location | 4,023,763 – 4,023,883 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -40.06 |
| Consensus MFE | -31.54 |
| Energy contribution | -31.58 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4023763 120 + 23771897 AUGCCAGUAAACAAAAUUUCGAGAGGGCUCCAGUGGCGAGGUUACCAGAUAUUCGCGAAUGCGUUUGUGUGGGUAGCAGUGUUGCGAAACAUGUGUUUCCCAUGGAGAUCUGGCAGGCCA .((((((..........(((....)))((((((..(((..(((((((.(((..(((....)))..))).).))))))..)))..)(((((....)))))...)))))..))))))..... ( -43.60) >DroSec_CAF1 3301 120 + 1 AUGCCAGUAAACAAAAUUUCGAGAGGGCUCCAGUGGCGAGGUUGCCAGAUAGUCGCGAAUGCGUGUGUGUGGGUAGCAGUGUUGCGAAGCAUGGUUUUUCCAUGGAGAUCUGGCAGGCCA .((((((..........(((....)))((((.(..(((..((((((..(((..((((....))))..))).))))))..)))..).....((((.....))))))))..))))))..... ( -40.80) >DroSim_CAF1 3343 120 + 1 AUGCCAGUAAACAAAAUUUCGAGAGGGCUCCAGUGGCGAGGUUGCCAGAUAGUCGCGAAUGCGUGUGUGUGGGUAGCAGUGUUGCGAAGCAUGGUUUUUCCAUGGAGAUCUGGCAGGCCA .((((((..........(((....)))((((.(..(((..((((((..(((..((((....))))..))).))))))..)))..).....((((.....))))))))..))))))..... ( -40.80) >DroEre_CAF1 3308 117 + 1 AUACCAGUAAACAAAAUUUCGAGAGGGA-CCAGUAGCGAGGUUGCCAGAUGUUCGCGACUG--UAUGUGUGGGUGGUAGUGUUGCGAAGCGUGGGUUUCCCACGGACAUCUGGCAGGCCA ................(..(....)..)-..........((((((((((((((((((((..--(((.........)))..)))))))..((((((...)))))).)))))))))).))). ( -40.90) >DroYak_CAF1 3239 118 + 1 AUGCCAGUAAACAAAAUUUCGAGAGGGCUCCAGCAGCGAGGUUACCAGAUGUUCGCGAAUG--UGUGUGUGGGUGGUAGUGUUGCGAAACGUGGGUUUUCCAUGGAGAUGUGGCAGGCCA .(((((...........(((....)))((((.((((((..(..(((..(..(.(((....)--)).)..).)))..)..)))))).....((((.....))))))))...)))))..... ( -34.20) >consensus AUGCCAGUAAACAAAAUUUCGAGAGGGCUCCAGUGGCGAGGUUGCCAGAUAUUCGCGAAUGCGUGUGUGUGGGUAGCAGUGUUGCGAAGCAUGGGUUUUCCAUGGAGAUCUGGCAGGCCA .((((((..........(((....)))((((.((((((..(((((((.(((..(((....)))..))).).))))))..)))))).....((((.....))))))))..))))))..... (-31.54 = -31.58 + 0.04)

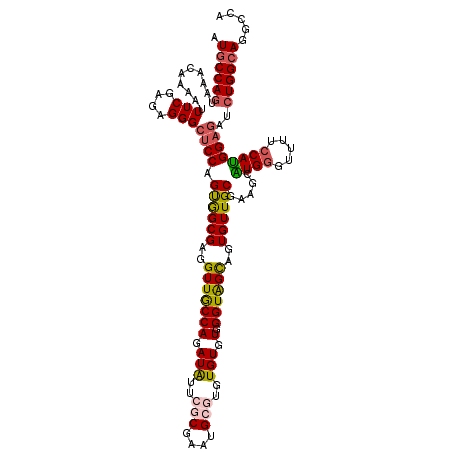
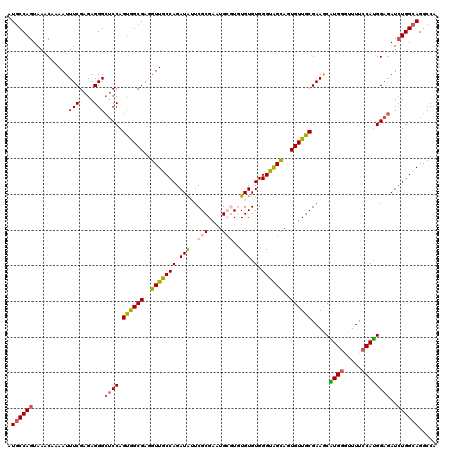
| Location | 4,023,763 – 4,023,883 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -22.87 |
| Energy contribution | -22.83 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4023763 120 - 23771897 UGGCCUGCCAGAUCUCCAUGGGAAACACAUGUUUCGCAACACUGCUACCCACACAAACGCAUUCGCGAAUAUCUGGUAACCUCGCCACUGGAGCCCUCUCGAAAUUUUGUUUACUGGCAU .((..((((((((.....((((((((....)))))(((....)))...)))......(((....)))...)))))))).))..((((...((((..............))))..)))).. ( -30.44) >DroSec_CAF1 3301 120 - 1 UGGCCUGCCAGAUCUCCAUGGAAAAACCAUGCUUCGCAACACUGCUACCCACACACACGCAUUCGCGACUAUCUGGCAACCUCGCCACUGGAGCCCUCUCGAAAUUUUGUUUACUGGCAU .((..((((((((...(((((.....)))))....(((....)))............(((....)))...)))))))).))..((((...((((..............))))..)))).. ( -29.84) >DroSim_CAF1 3343 120 - 1 UGGCCUGCCAGAUCUCCAUGGAAAAACCAUGCUUCGCAACACUGCUACCCACACACACGCAUUCGCGACUAUCUGGCAACCUCGCCACUGGAGCCCUCUCGAAAUUUUGUUUACUGGCAU .((..((((((((...(((((.....)))))....(((....)))............(((....)))...)))))))).))..((((...((((..............))))..)))).. ( -29.84) >DroEre_CAF1 3308 117 - 1 UGGCCUGCCAGAUGUCCGUGGGAAACCCACGCUUCGCAACACUACCACCCACACAUA--CAGUCGCGAACAUCUGGCAACCUCGCUACUGG-UCCCUCUCGAAAUUUUGUUUACUGGUAU .((..((((((((((.((((((...))))))..((((.((.................--..)).)))))))))))))).))....(((..(-(......(((....)))...))..))). ( -34.11) >DroYak_CAF1 3239 118 - 1 UGGCCUGCCACAUCUCCAUGGAAAACCCACGUUUCGCAACACUACCACCCACACACA--CAUUCGCGAACAUCUGGUAACCUCGCUGCUGGAGCCCUCUCGAAAUUUUGUUUACUGGCAU .....(((((........(((.....)))............................--..((((.((...((..(((.......)))..))....)).))))...........))))). ( -18.60) >consensus UGGCCUGCCAGAUCUCCAUGGAAAACCCAUGCUUCGCAACACUGCUACCCACACACACGCAUUCGCGAAUAUCUGGCAACCUCGCCACUGGAGCCCUCUCGAAAUUUUGUUUACUGGCAU ..(((((((((((...(((((.....)))))..((((...........................))))..)))))))).((........))........................))).. (-22.87 = -22.83 + -0.04)



| Location | 4,023,803 – 4,023,923 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -34.49 |
| Consensus MFE | -22.98 |
| Energy contribution | -22.86 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4023803 120 + 23771897 GUUACCAGAUAUUCGCGAAUGCGUUUGUGUGGGUAGCAGUGUUGCGAAACAUGUGUUUCCCAUGGAGAUCUGGCAGGCCAGUGACACUAUGAUGGUAACUUUUAAAAUGGUAAAGUCAAG (((((((.(((..(((....)))..))).).))))))(((((..((((((....)))))...........(((....))))..))))).((((..((.((........))))..)))).. ( -33.60) >DroSec_CAF1 3341 116 + 1 GUUGCCAGAUAGUCGCGAAUGCGUGUGUGUGGGUAGCAGUGUUGCGAAGCAUGGUUUUUCCAUGGAGAUCUGGCAGGCCAGUGACACUAUGUUGGUAACUUUUAAAAUUGUAAAAU---- ((((((((((((((((((.(((.............)))...))))))..(((((.....))))).....((((....)))).....)))).)))))))).................---- ( -33.32) >DroSim_CAF1 3383 120 + 1 GUUGCCAGAUAGUCGCGAAUGCGUGUGUGUGGGUAGCAGUGUUGCGAAGCAUGGUUUUUCCAUGGAGAUCUGGCAGGCCAGUGACACUAUGUUGGUAGCCUUUAAAAUUGUAAAAUCAAA ..((((((((..((((((.(((.............)))...))))))..(((((.....)))))...))))))))(((.(.((((.....)))).).))).................... ( -36.22) >DroEre_CAF1 3347 104 + 1 GUUGCCAGAUGUUCGCGACUG--UAUGUGUGGGUGGUAGUGUUGCGAAGCGUGGGUUUCCCACGGACAUCUGGCAGGCCAGUAGCAGU------------U-AAAAAUUGUACACAU-CU .((((((((((((((((((..--(((.........)))..)))))))..((((((...)))))).)))))))))))....(((.((((------------.-....)))))))....-.. ( -37.70) >DroYak_CAF1 3279 106 + 1 GUUACCAGAUGUUCGCGAAUG--UGUGUGUGGGUGGUAGUGUUGCGAAACGUGGGUUUUCCAUGGAGAUGUGGCAGGCCAGUCGCAGU------------UUUUAAAUUUUAGAAACAAU .....((.(((((....))))--).))(((((.((((..(((..((...(((((.....)))))....))..))).)))).)))))((------------(((((.....)))))))... ( -31.60) >consensus GUUGCCAGAUAUUCGCGAAUGCGUGUGUGUGGGUAGCAGUGUUGCGAAGCAUGGGUUUUCCAUGGAGAUCUGGCAGGCCAGUGACACUAUG_UGGUA_C_UUUAAAAUUGUAAAAUCAAU (((((((.(((..(((....)))..))).).))))))((((((((....(((((.....)))))......(((....)))))))))))................................ (-22.98 = -22.86 + -0.12)

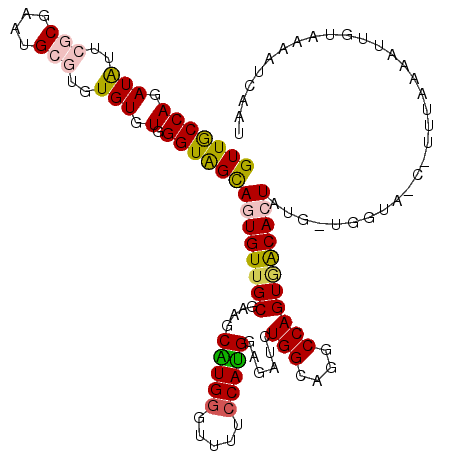
| Location | 4,023,803 – 4,023,923 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.15 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4023803 120 - 23771897 CUUGACUUUACCAUUUUAAAAGUUACCAUCAUAGUGUCACUGGCCUGCCAGAUCUCCAUGGGAAACACAUGUUUCGCAACACUGCUACCCACACAAACGCAUUCGCGAAUAUCUGGUAAC .....................(((((((...((((((..((((....)))).......((.(((((....))))).)))))))).............(((....)))......))))))) ( -29.20) >DroSec_CAF1 3341 116 - 1 ----AUUUUACAAUUUUAAAAGUUACCAACAUAGUGUCACUGGCCUGCCAGAUCUCCAUGGAAAAACCAUGCUUCGCAACACUGCUACCCACACACACGCAUUCGCGACUAUCUGGCAAC ----................(((.(((......).)).)))....((((((((...(((((.....)))))....(((....)))............(((....)))...)))))))).. ( -22.60) >DroSim_CAF1 3383 120 - 1 UUUGAUUUUACAAUUUUAAAGGCUACCAACAUAGUGUCACUGGCCUGCCAGAUCUCCAUGGAAAAACCAUGCUUCGCAACACUGCUACCCACACACACGCAUUCGCGACUAUCUGGCAAC ...................((((((...((.....))...))))))(((((((...(((((.....)))))....(((....)))............(((....)))...)))))))... ( -28.30) >DroEre_CAF1 3347 104 - 1 AG-AUGUGUACAAUUUUU-A------------ACUGCUACUGGCCUGCCAGAUGUCCGUGGGAAACCCACGCUUCGCAACACUACCACCCACACAUA--CAGUCGCGAACAUCUGGCAAC ..-...............-.------------...((.....)).((((((((((.((((((...))))))..((((.((.................--..)).)))))))))))))).. ( -29.31) >DroYak_CAF1 3279 106 - 1 AUUGUUUCUAAAAUUUAAAA------------ACUGCGACUGGCCUGCCACAUCUCCAUGGAAAACCCACGUUUCGCAACACUACCACCCACACACA--CAUUCGCGAACAUCUGGUAAC ....................------------..(((((.(((....)))........(((.....)))....)))))....(((((......(.(.--.....).)......))))).. ( -12.60) >consensus AUUGAUUUUACAAUUUUAAA_G_UACCA_CAUAGUGUCACUGGCCUGCCAGAUCUCCAUGGAAAACCCAUGCUUCGCAACACUGCUACCCACACACACGCAUUCGCGAAUAUCUGGCAAC .............................................((((((((...(((((.....)))))..((((...........................))))..)))))))).. (-17.03 = -17.15 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:10 2006