
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,015,304 – 4,015,408 |
| Length | 104 |
| Max. P | 0.960243 |

| Location | 4,015,304 – 4,015,408 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -28.49 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4015304 104 - 23771897 UUUUGCACCUCAGAUCCAGCCAUU-UGGUUGAUGAGCACCAGUUCGUAGACCAUCAAAGUGGCAGCCAUCUGUCCAGAUAUUGAGUUGCU---UAAAAUGCACUAG-AA ....(((.(((((.....((((((-(((((.((((((....)))))).)))))....))))))....((((....)))).))))).))).---.............-.. ( -30.00) >DroSec_CAF1 5012 100 - 1 UUCUGCACCUCAGAUCCAGCCAUU-UGGUUGAUGAGCACCAGUUCGUAGACCAUCAAAGUGGCAGCCAUCUGAUCAGAUAUA----UG-U---UCAUAUAUCCAGAAAG (((((....((((((...((((((-(((((.((((((....)))))).)))))....))))))....))))))...((((((----(.-.---..)))))))))))).. ( -33.30) >DroSim_CAF1 5785 101 - 1 UUCUGCACCUCAGAUCCAGCCAUUUUGGUUGAUGAGCACCAGUUCGUAGACCAUCAAAGUGGCAGCCAUCUGAUCAGAUAUA----UG-U---UCAUAUGUCCAGGAAA (((((....((((((...((((((((((((.((((((....)))))).)))))...)))))))....))))))...((((((----(.-.---..)))))))))))).. ( -34.10) >DroEre_CAF1 6744 104 - 1 CCUUGCACCUCGGAUCCAGCCAUU-UGGUUAAUGAGCACCAGUUCAUAGACCAUCAAAGUAGCAGCCAUCUAUCCGGAUAUUGAGUAGCU---UAAAAUGCAGU-AGAA ..((((..(((((((((.......-(((((.((((((....)))))).))))).....((((.......))))..)))).)))))..((.---......)).))-)).. ( -23.60) >DroYak_CAF1 5066 104 - 1 UUUUGCACCUCGGAUCCAGCCAUU-UGGUUAAUGAGCACCAGUUCAUAGACCAUCAGAGUAGCAGCCAUCUAUCCACAUAUUGAGUUGCU---UAAAAUGCAAU-AAAA ..(((((.(((((......))...-(((((.((((((....)))))).)))))...))).((((((((.............)).))))))---.....))))).-.... ( -26.92) >DroMoj_CAF1 9624 98 - 1 GUCUGCACCUCGGAGCCACUCAUU-UGAUUGAUUAGCACCAGCUCGUAGACCAUCAGCGUGGCGGCCAUCUGCAA-GAG--GGAGAGGAUAAA---UAAGGGCC----A .((((.....))))(((((.(...-((.((.((.(((....))).)).)).))...).)))))((((.(((.(..-...--).))).......---....))))----. ( -23.00) >consensus UUCUGCACCUCAGAUCCAGCCAUU_UGGUUGAUGAGCACCAGUUCGUAGACCAUCAAAGUGGCAGCCAUCUGUCCAGAUAUUGAGUUGCU___UAAAAUGCACU_AAAA ..........(((((...((((((.(((((.((((((....)))))).)))))....))))))....)))))..................................... (-17.83 = -18.58 + 0.75)

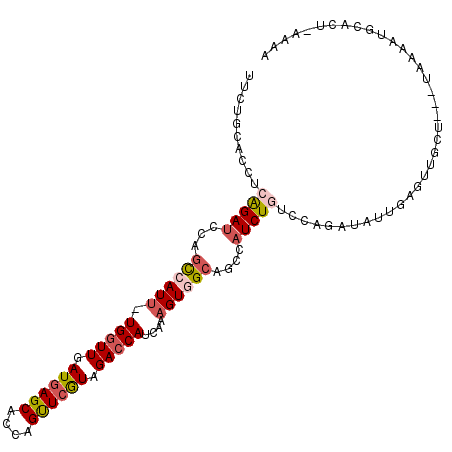
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:02 2006