
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,014,914 – 4,015,017 |
| Length | 103 |
| Max. P | 0.528284 |

| Location | 4,014,914 – 4,015,017 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.31 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -11.79 |
| Energy contribution | -12.27 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
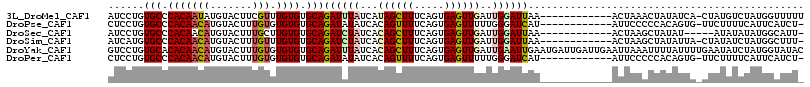
>3L_DroMel_CAF1 4014914 103 + 23771897 AUCCUGUGCCCACAAUAUGUACUUCGUUUGUGUGCAGAUUUAUCAUAGCUUUCAGUGAGUUGAUUGGAUUAA------------ACUAAACUAUAUCA-CUAUGUCUAUGGUUUUU ....(.(((.(((((.(((.....)))))))).))).)...((((((((....(((((...(.((((.....------------.)))).)....)))-))..).))))))).... ( -20.40) >DroPse_CAF1 4374 102 + 1 CUCCUGUGCCCACAACAUGUACUUUGUGUGUGUGCAGAUAUAUCACAGUUUUCAGUGAGUUUUUGGGAUCAU------------AUUCCCCCACAGUG-UUCUUUUCAUUCAUCU- ...(((..(.((((.((.......)))))).)..))).....((((........))))......((((....------------..))))........-................- ( -20.80) >DroSec_CAF1 4634 98 + 1 AUCCUGUGCCCACAACAUGUACUUUGCUUGUGUGCAGAUCUAUCACAGCUUUCAGUGAGUUGAUUGGAUUAA------------ACUAAGCUAUAU-----AUAUAUAUGGCAUU- ......(((.(((((((.......)).))))).)))((((((...((((((.....))))))..))))))..------------.....(((((((-----....)))))))...- ( -25.20) >DroSim_CAF1 5397 102 + 1 AUCAUGUGCCCACAACAUGUACUUUGUUUGUGUGCAGAUCUAUCACAGCUUUCAGUGAGUUGAUUGGAUUAA------------ACUAAGCUAUAUUA-CUAUAUCUAUGGCUUU- ......(((.(((((((.......)).))))).)))((((((...((((((.....))))))..))))))..------------...((((((((...-.......)))))))).- ( -23.90) >DroYak_CAF1 4647 116 + 1 GUCCUGUGCACACAACAUGUACUUUGUGUGUGUGCAGAUUUAUCACAGCUUUCAGUGAGUUGAUUGAAUUGAAUGAUUGAUUGAAUUAAAUUUUAUUUUGAAUAUCUAUGGUAUAC ...(((..((((((.((.......))))))))..)))...(((((.((..((((((((.((((((.(((..(....)..))).))))))...))))..))))...)).)))))... ( -29.10) >DroPer_CAF1 4368 102 + 1 CUCCUGUGCCCACAACAUGUACUUUGUGUGUGUGCAGAUAUAUCACAGUUUUCAGUGAGUUUUUGGGAUCAU------------AUUCCCCCACAGUG-UUCUUUUCAUUCAUCU- ...(((..(.((((.((.......)))))).)..))).....((((........))))......((((....------------..))))........-................- ( -20.80) >consensus AUCCUGUGCCCACAACAUGUACUUUGUGUGUGUGCAGAUAUAUCACAGCUUUCAGUGAGUUGAUUGGAUUAA____________ACUAAACUAUAUUA_CUAUAUCUAUGGAUUU_ ......(((.(((((((.......))).)))).)))((((((...((((((.....))))))..)))))).............................................. (-11.79 = -12.27 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:01 2006