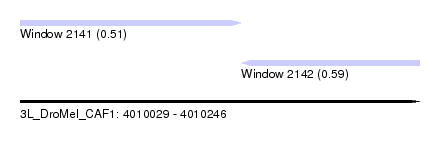
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,010,029 – 4,010,246 |
| Length | 217 |
| Max. P | 0.594691 |

| Location | 4,010,029 – 4,010,149 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -29.01 |
| Energy contribution | -29.79 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.94 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4010029 120 + 23771897 CUCACAUGUAUCUCGGCAGCUAAUCGAAAUGCAAUCUGACGGAUGCAUCCGAAUCCUUCGGUCUGCUGCUGCAGAGUGAACCCACUUUAUUCCAAUUCCCACAGAAGUGUCUCAUAAUUG ..........((((((((((..(((((((((((.((.....)))))))........))))))..))))))).)))((((...((((((...............))))))..))))..... ( -30.56) >DroSec_CAF1 4747 119 + 1 CUCGCAUGUAUCUCAGCCGCUAAUCGAAAUGCAAUCUGACGGAUGCAUGCUAAUCCUUCGGUCAGCUGCUGCAGAGUGAACUCACUUU-UUCCAAUUCUCACAGAAGUGUCUCAUAAUUG ..........(((((((.(((.(((((((((((.((.....)))))))........)))))).))).)))).)))((((...((((((-(............)))))))..))))..... ( -28.90) >DroSim_CAF1 4774 119 + 1 CUCGCAUGUAUCUCAGCAGCUAAUCGAACUGCAAUCUGACGGAUGCAUCCGAAUCCUUCGGUCAGCUGCUGCAGAGUGAACUCACUUU-UUGCAAUUCCCACAGAAGUGUCUCAUAAUUG ..........(((((((((((.(((((((........).((((....)))).....)))))).)))))))).)))((((...((((((-(((.......)).)))))))..))))..... ( -33.10) >consensus CUCGCAUGUAUCUCAGCAGCUAAUCGAAAUGCAAUCUGACGGAUGCAUCCGAAUCCUUCGGUCAGCUGCUGCAGAGUGAACUCACUUU_UUCCAAUUCCCACAGAAGUGUCUCAUAAUUG ..........(((((((((((.(((((((((((.((.....)))))))........)))))).)))))))).)))((((...((((((...............))))))..))))..... (-29.01 = -29.79 + 0.78)


| Location | 4,010,149 – 4,010,246 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.24 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -24.29 |
| Energy contribution | -25.35 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4010149 97 - 23771897 CUUGUCAUGGGUGGAGUUGAGCCCUUCACAAUG-----------------GUGCCCGUCUGAAGUGCCACUAAUUAGGGGCUUUGGACCUAAGCUAGUACCAGGAUGUAAACAA ...(((.(((((((((.......))))))..((-----------------((....(((..((((.((........)).))))..)))....))))...))).)))........ ( -29.80) >DroSec_CAF1 4866 96 - 1 CUUGUCAUGGGCGGAGUUGAGUCGUCCACAAUG-----------------GUGUCCGUCCGAAGUGCCACUAAUUAGGGGCUUUGGAC-UGAGCUAGUCCCAGGAUGUAAACAA ...(((.(((((..((((.((.((..(((....-----------------)))..))((((((((.((........)).)))))))))-).)))).).)))).)))........ ( -32.90) >DroSim_CAF1 4893 96 - 1 CUUGUCAUGGGUGGAGUUGAGUCCUCCACAAUG-----------------GUGUCCGUCCGAAGUGCCACUAAUUAGGGGCUUUGGAC-UGAGCUAGUCCCAGGAUGUAAACAA ...(((.(((((((((.......))))))..((-----------------((.((.(((((((((.((........)).)))))))))-.))))))...))).)))........ ( -35.70) >DroYak_CAF1 5613 113 - 1 UUUGUCAGGGGCGGAGUUGAGCCCUCCACAAUGGAUUUGGUUGUACUAGGGUGUCCGUCUGAAGUGCCACUAAUUAGGGGCUUUGGAC-UGAGCUAGUCCGAGUAUGUAAACAA .((((..(((((........)))))..))))((((((.((((......((....))(((..((((.((........)).))))..)))-..))))))))))............. ( -36.10) >consensus CUUGUCAUGGGCGGAGUUGAGCCCUCCACAAUG_________________GUGUCCGUCCGAAGUGCCACUAAUUAGGGGCUUUGGAC_UGAGCUAGUCCCAGGAUGUAAACAA ...(((.(((((((((.......)))))).....................((....(((((((((.((........)).)))))))))....)).....))).)))........ (-24.29 = -25.35 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:59 2006