
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,998,163 – 3,998,256 |
| Length | 93 |
| Max. P | 0.562029 |

| Location | 3,998,163 – 3,998,256 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -19.03 |
| Energy contribution | -19.23 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
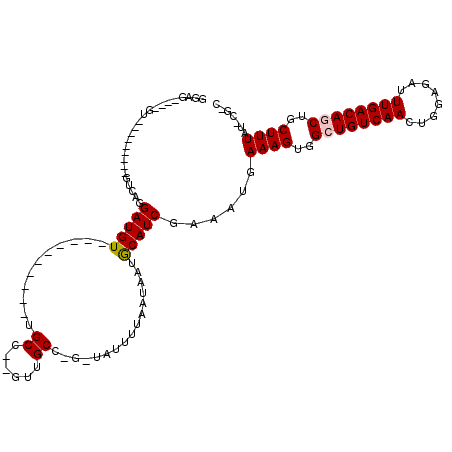
>3L_DroMel_CAF1 3998163 93 + 23771897 GGAG----UU---------GUCAGGAUGU----------UGCC--ACUGCCUG-UAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAUGCC-G ((((----(.---------(.(((....)----------))).--))).))((-((((.....))))))........((((..((((((((........))))))))..)))).....-. ( -25.00) >DroVir_CAF1 64273 93 + 1 GGAU----GU---------GGCAGGAUGU----------UGCCUUGUUGCC-G-CAUUUUAAUAAUACAUCGAAAUGAAAGUGGCUGUCAACUAGAGAUUUGACAUCUGCUUUGU-GG-C ((((----((---------(((((.(...----------.....).)))))-)-)))))..................((((..(.((((((........)))))).)..))))..-..-. ( -25.90) >DroGri_CAF1 80378 111 + 1 GGAG----GUGGGUACUUUGCCAGGAUGU--UGCCUUGUUGCCUUGUUGCC-GUUAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAUCUGCUUUGU-CG-C .(((----((.(((.....))).((((((--(.((..((((.(..((..(.-.((((((..((.....))..))))))..)..)).).)))).))......))))))))))))..-..-. ( -25.60) >DroMoj_CAF1 73015 99 + 1 GGAU----GU---------GCCAGGAUGUGUUGC----UUGCCUUGUUGCC-G-CAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUAGAGAUUUGACAGCUGCUUUGU-CG-C ((..----..---------((.(((..(.....)----...))).))..))-(-((((.....)))))..(((....((((..((((((((........))))))))..)))).)-))-. ( -30.50) >DroAna_CAF1 80015 100 + 1 GGAG----GUG---UCGGUGUCAGGAUGU----------UGCC--AGUGCCUG-UAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAGCCAAC ...(----((.---(((((((((((.((.----------...)--)...))))-............)))))))....((((..((((((((........))))))))..)))).)))... ( -33.30) >DroPer_CAF1 72353 95 + 1 GGAGGAUGGU---------GUCUGGAUGU----------UGCC--GCUGCC-G-UAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUA-GCU-U ((.((.((((---------(.(.....).----------))))--))).))-(-((((.....))))).........((((..((((((((........))))))))..)))).-...-. ( -28.30) >consensus GGAG____GU_________GUCAGGAUGU__________UGCC__GUUGCC_G_UAUUUUAAUAAUGCAUCGAAAUGAAAGUGGCUGUCAACUGGAGAUUUGACAGCUGCUUUAU_CG_C ........................(((((...........((......))................)))))......((((..((((((((........))))))))..))))....... (-19.03 = -19.23 + 0.20)

| Location | 3,998,163 – 3,998,256 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -12.26 |
| Energy contribution | -12.23 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3998163 93 - 23771897 C-GGCAUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUA-CAGGCAGU--GGCA----------ACAUCCUGAC---------AA----CUCC .-.((((((((..((((((((........))))))))..)))).......))))............-((((..((--(...----------.)))))))..---------..----.... ( -24.11) >DroVir_CAF1 64273 93 - 1 G-CC-ACAAAGCAGAUGUCAAAUCUCUAGUUGACAGCCACUUUCAUUUCGAUGUAUUAUUAAAAUG-C-GGCAACAAGGCA----------ACAUCCUGCC---------AC----AUCC (-((-..((((....((((((........))))))....)))).........(((((.....))))-)-))).....((((----------......))))---------..----.... ( -20.00) >DroGri_CAF1 80378 111 - 1 G-CG-ACAAAGCAGAUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUAAC-GGCAACAAGGCAACAAGGCA--ACAUCCUGGCAAAGUACCCAC----CUCC (-(.-.....))...((((((........))))))((((...((.....))(((..............-(....)..(....)...)))--......))))...........----.... ( -17.40) >DroMoj_CAF1 73015 99 - 1 G-CG-ACAAAGCAGCUGUCAAAUCUCUAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUG-C-GGCAACAAGGCAA----GCAACACAUCCUGGC---------AC----AUCC .-((-(.((((..((((((((........))))))))..))))....))).((((((.....))))-)-)((....(((...----.........))).))---------..----.... ( -21.10) >DroAna_CAF1 80015 100 - 1 GUUGGCUAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUA-CAGGCACU--GGCA----------ACAUCCUGACACCGA---CAC----CUCC (((((.....(((((((((((........)))))))).....((.....)))))............-((((...(--(...----------.)).))))...))))---)..----.... ( -26.20) >DroPer_CAF1 72353 95 - 1 A-AGC-UAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUA-C-GGCAGC--GGCA----------ACAUCCAGAC---------ACCAUCCUCC .-.((-.((((..((((((((........))))))))..)))).....((.(((..(((....)))-.-.))).)--))).----------..........---------.......... ( -18.10) >consensus G_CG_ACAAAGCAGCUGUCAAAUCUCCAGUUGACAGCCACUUUCAUUUCGAUGCAUUAUUAAAAUA_C_GGCAAC__GGCA__________ACAUCCUGAC_________AC____CUCC ..........((.((((((((........))))))))..............(((..(((....)))....))).....))........................................ (-12.26 = -12.23 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:56 2006