
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,959,252 – 3,959,419 |
| Length | 167 |
| Max. P | 0.989578 |

| Location | 3,959,252 – 3,959,350 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.11 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -15.34 |
| Energy contribution | -15.87 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
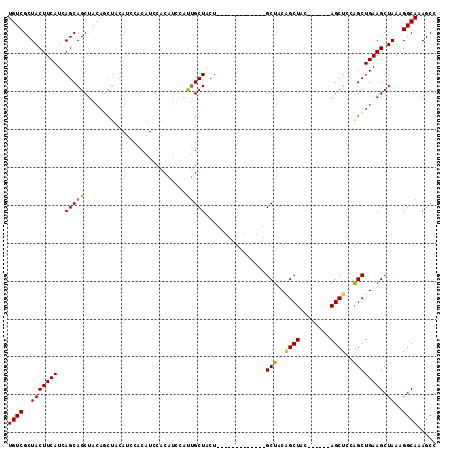
>3L_DroMel_CAF1 3959252 98 + 23771897 AGUAGCACUCAAAGUGAGUUUGCGACGCAUUGCCGCAUUGUCGCUACUUCAUCAGCAGCUA-C------AGCUACAUCCACAUCCACAUCCAUUGCUACU-------------GCUAC (((((((......((((((..(((((((......))...))))).)))......(.(((..-.------.))).).........)))......)))))))-------------..... ( -22.50) >DroPse_CAF1 32372 99 + 1 AGUAGCACUCAAAGUGAGUUUGCGACGCAUUGCCGCAUUGUCGCUACUUCAUCAGCAGGCAACGGGAGUGGCAACGGCGACGGCGACAACGACGGCAAC------------------- .((((.((((.....))))))))......(((((((.(((((((((((((.......(....).))))))))..((....))..))))).).)))))).------------------- ( -36.60) >DroSim_CAF1 35015 98 + 1 AGUAGCACUCAAAGUGAGUUUGCGACGCAUUGCCGCAUUGUCGCUACUUCAUCAGCAGCUA-C------AGCUACAUCCACAUCCACAUCCAUUGCUACU-------------GCUAC (((((((......((((((..(((((((......))...))))).)))......(.(((..-.------.))).).........)))......)))))))-------------..... ( -22.50) >DroEre_CAF1 32625 111 + 1 AGUAGCACUCAACGUGAGUUUGCGACGCAUUGCCGCAUUGUCGCUACUUCAUCAGCAGCUA-C------AACUACAUCCACAUCCACAUCCACUGCUACUUGCUGUAGCUGCAGCUCC ((((((((((.....))))..((((.((......)).)))).))))))......(((((((-(------(........................((.....)))))))))))...... ( -28.35) >DroYak_CAF1 33846 98 + 1 AGUAGCACUCAACGUGAGUUUGCGACGCAUUGCCGCAUUGUCGCUACUUCAUCAGCAGCUA-------------CAUCCACAUCCACAUCCACAGCUAC-------AACUGCAGCUCC .(((((((((.....))))..(((((((......))...))))).............))))-------------)..................((((.(-------....).)))).. ( -21.50) >DroAna_CAF1 33282 86 + 1 AGUAGCACUCAAAGUGAGUUUGCGACGCAUUGCCGCAUUGUCGCUACUUCAUCAGCA---A-C------AGCUACAGCCCCGGCGAAAGCUUCUGU---------------------- .(((((((((.....))))..(((((((......))...))))).............---.-.------.)))))......(((....))).....---------------------- ( -23.90) >consensus AGUAGCACUCAAAGUGAGUUUGCGACGCAUUGCCGCAUUGUCGCUACUUCAUCAGCAGCUA_C______AGCUACAUCCACAUCCACAUCCACUGCUAC______________GCU_C .(((((((((.....))))..(((((((......))...)))))..................................................)))))................... (-15.34 = -15.87 + 0.53)

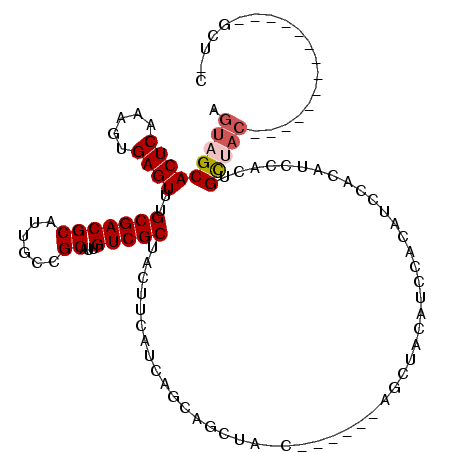

| Location | 3,959,252 – 3,959,350 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.11 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -21.40 |
| Energy contribution | -22.73 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3959252 98 - 23771897 GUAGC-------------AGUAGCAAUGGAUGUGGAUGUGGAUGUAGCU------G-UAGCUGCUGAUGAAGUAGCGACAAUGCGGCAAUGCGUCGCAAACUCACUUUGAGUGCUACU .....-------------(((((((.(.((.((((...(((((((((((------(-((((((((.....)))))).....))))))..)))))).))...)))).)).).))))))) ( -35.20) >DroPse_CAF1 32372 99 - 1 -------------------GUUGCCGUCGUUGUCGCCGUCGCCGUUGCCACUCCCGUUGCCUGCUGAUGAAGUAGCGACAAUGCGGCAAUGCGUCGCAAACUCACUUUGAGUGCUACU -------------------(((((((.(((((((((((((((....((.((....)).))..)).)))).....))))))))))))))))..((.((..((((.....)))))).)). ( -35.60) >DroSim_CAF1 35015 98 - 1 GUAGC-------------AGUAGCAAUGGAUGUGGAUGUGGAUGUAGCU------G-UAGCUGCUGAUGAAGUAGCGACAAUGCGGCAAUGCGUCGCAAACUCACUUUGAGUGCUACU .....-------------(((((((.(.((.((((...(((((((((((------(-((((((((.....)))))).....))))))..)))))).))...)))).)).).))))))) ( -35.20) >DroEre_CAF1 32625 111 - 1 GGAGCUGCAGCUACAGCAAGUAGCAGUGGAUGUGGAUGUGGAUGUAGUU------G-UAGCUGCUGAUGAAGUAGCGACAAUGCGGCAAUGCGUCGCAAACUCACGUUGAGUGCUACU ...((((......)))).(((((((.(.(((((((...........(((------(-(.((((((.....)))))).)))))(((((.....)))))....))))))).).))))))) ( -45.00) >DroYak_CAF1 33846 98 - 1 GGAGCUGCAGUU-------GUAGCUGUGGAUGUGGAUGUGGAUG-------------UAGCUGCUGAUGAAGUAGCGACAAUGCGGCAAUGCGUCGCAAACUCACGUUGAGUGCUACU ..((((((....-------))))))((((...(.(((((((.((-------------(.((((((.....)))))).))).((((((.....))))))...))))))).)...)))). ( -38.00) >DroAna_CAF1 33282 86 - 1 ----------------------ACAGAAGCUUUCGCCGGGGCUGUAGCU------G-U---UGCUGAUGAAGUAGCGACAAUGCGGCAAUGCGUCGCAAACUCACUUUGAGUGCUACU ----------------------.....(((((......)))))((((((------(-(---(((((......)))))))).((((((.....)))))).((((.....))))))))). ( -30.80) >consensus G_AGC______________GUAGCAGUGGAUGUGGAUGUGGAUGUAGCU______G_UAGCUGCUGAUGAAGUAGCGACAAUGCGGCAAUGCGUCGCAAACUCACUUUGAGUGCUACU ...................(((((...................................((((((.....)))))).....((((((.....)))))).((((.....))))))))). (-21.40 = -22.73 + 1.33)

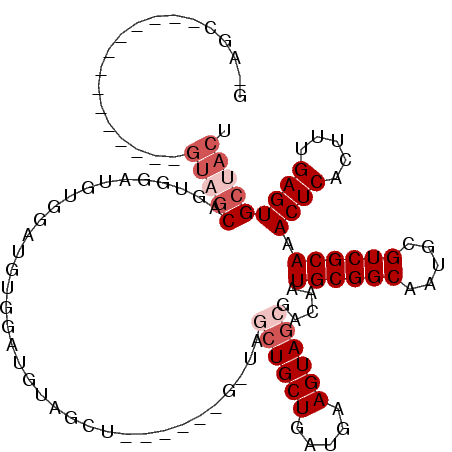

| Location | 3,959,290 – 3,959,384 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -15.69 |
| Energy contribution | -15.57 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3959290 94 + 23771897 UGUCGCUACUUCAUCAGCAGCUACAGCUACAUCCACAUCCACAUCCAUUGCUACU-------------GCUACGGCUAC------AGCUCCAGCUGAAGCUAAAGGCAAAGCC (((((((........)))((((.(((((.....................(((...-------------((....))...------)))...))))).))))...))))..... ( -21.75) >DroSec_CAF1 31766 94 + 1 UGUCGCUACUUCAUCAGCAGCUACAGCUACAUCCACAUCCACAUCCAUUGCUACU-------------GCUACAGCUAC------AGCUCCAGCUGAAGCUAAAGGCAAAGCC (((.(((.((.....)).))).)))........................(((..(-------------(((..((((.(------(((....)))).))))...)))).))). ( -21.20) >DroSim_CAF1 35053 94 + 1 UGUCGCUACUUCAUCAGCAGCUACAGCUACAUCCACAUCCACAUCCAUUGCUACU-------------GCUACAGCUAC------AGCUCCGGCUGAAGCUAAAGGCAAAGCC (((.(((.((.....)).))).)))........................(((..(-------------(((..((((.(------(((....)))).))))...)))).))). ( -21.30) >DroEre_CAF1 32663 107 + 1 UGUCGCUACUUCAUCAGCAGCUACAACUACAUCCACAUCCACAUCCACUGCUACUUGCUGUAGCUGCAGCUCCUGCUCC------AGCUCCAGCUGAAGCUAAAGGCAAAGCC ((((..(((((((..(((((...........................)))))....((((.(((((.(((....))).)------)))).))))))))).))..))))..... ( -30.23) >DroYak_CAF1 33884 100 + 1 UGUCGCUACUUCAUCAGCAGCUA------CAUCCACAUCCACAUCCACAGCUAC-------AACUGCAGCUCCAGCUGAAGGAGAAGCUGAAGCUGAAGCUAAAGGCAAAGCC ((((..(((((((.(..(((((.------..(((.............(((....-------..)))((((....))))..)))..)))))..).))))).))..))))..... ( -27.10) >consensus UGUCGCUACUUCAUCAGCAGCUACAGCUACAUCCACAUCCACAUCCAUUGCUACU_____________GCUACAGCUAC______AGCUCCAGCUGAAGCUAAAGGCAAAGCC ((((..(((((((..(((((...........................)))))................(((..((((........))))..)))))))).))..))))..... (-15.69 = -15.57 + -0.12)

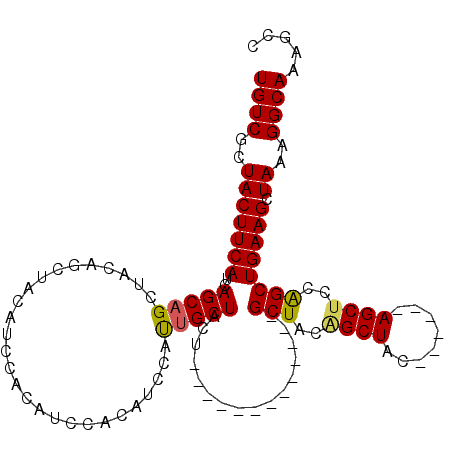
| Location | 3,959,290 – 3,959,384 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -19.93 |
| Energy contribution | -20.53 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3959290 94 - 23771897 GGCUUUGCCUUUAGCUUCAGCUGGAGCU------GUAGCCGUAGC-------------AGUAGCAAUGGAUGUGGAUGUGGAUGUAGCUGUAGCUGCUGAUGAAGUAGCGACA ((((..((.((((((....)))))))).------..))))((.((-------------(((.(((.((.((....)).))..))).))))).((((((.....)))))).)). ( -30.40) >DroSec_CAF1 31766 94 - 1 GGCUUUGCCUUUAGCUUCAGCUGGAGCU------GUAGCUGUAGC-------------AGUAGCAAUGGAUGUGGAUGUGGAUGUAGCUGUAGCUGCUGAUGAAGUAGCGACA ((((...((..(((((.((((....)))------).)))))..((-------------(...(((.....)))...)))))....))))((.((((((.....)))))).)). ( -30.50) >DroSim_CAF1 35053 94 - 1 GGCUUUGCCUUUAGCUUCAGCCGGAGCU------GUAGCUGUAGC-------------AGUAGCAAUGGAUGUGGAUGUGGAUGUAGCUGUAGCUGCUGAUGAAGUAGCGACA ((((...((..(((((.((((....)))------).)))))..((-------------(...(((.....)))...)))))....))))((.((((((.....)))))).)). ( -29.80) >DroEre_CAF1 32663 107 - 1 GGCUUUGCCUUUAGCUUCAGCUGGAGCU------GGAGCAGGAGCUGCAGCUACAGCAAGUAGCAGUGGAUGUGGAUGUGGAUGUAGUUGUAGCUGCUGAUGAAGUAGCGACA .(((....(((((((((((((....)))------)))))((.(((((((((((((.((....(((.....))).....))..))))))))))))).))..))))).))).... ( -45.20) >DroYak_CAF1 33884 100 - 1 GGCUUUGCCUUUAGCUUCAGCUUCAGCUUCUCCUUCAGCUGGAGCUGCAGUU-------GUAGCUGUGGAUGUGGAUGUGGAUG------UAGCUGCUGAUGAAGUAGCGACA .((((..(((.(((((.(((((.(((((((..(....)..))))))).))))-------).))))).))..)..)).))...((------(.((((((.....)))))).))) ( -36.10) >consensus GGCUUUGCCUUUAGCUUCAGCUGGAGCU______GUAGCUGUAGC_____________AGUAGCAAUGGAUGUGGAUGUGGAUGUAGCUGUAGCUGCUGAUGAAGUAGCGACA (((...)))....((((((((((.((((........)))).)))).............((((((.(..(..................)..).))))))..))))))....... (-19.93 = -20.53 + 0.60)

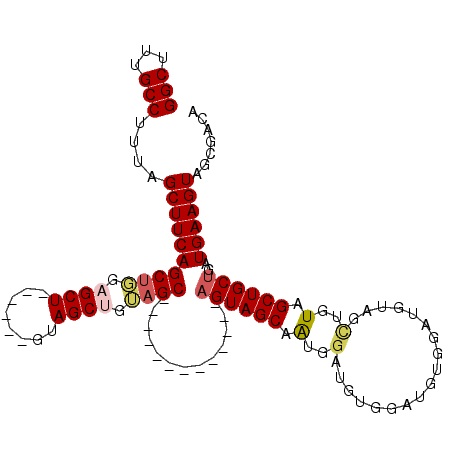

| Location | 3,959,323 – 3,959,419 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.66 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3959323 96 + 23771897 CACAUCCACAUCCAUUGCUACU-------------GCUACGGCUAC------AGCUCCAGCUGAAGCUAAAGGCAAAGCCAAAGCUUUAUAGCGAAGGCAUCAAGUGCAAGUAUC ..............((((.(((-------------(((..((((.(------(((....)))).))))...)))...(((...(((....)))...)))....)))))))..... ( -27.60) >DroSec_CAF1 31799 96 + 1 CACAUCCACAUCCAUUGCUACU-------------GCUACAGCUAC------AGCUCCAGCUGAAGCUAAAGGCAAAGCCAAAGCUUUAUAGCGAAGGCAUCAAGUGCAAGUAUC ..............((((.(((-------------(((..((((.(------(((....)))).))))...)))...(((...(((....)))...)))....)))))))..... ( -27.60) >DroSim_CAF1 35086 96 + 1 CACAUCCACAUCCAUUGCUACU-------------GCUACAGCUAC------AGCUCCGGCUGAAGCUAAAGGCAAAGCCAAAGCUUUAUAGCGAAGGCAUCAAGUGCAAGUAUC ..............((((.(((-------------(((..((((.(------(((....)))).))))...)))...(((...(((....)))...)))....)))))))..... ( -27.70) >DroEre_CAF1 32696 109 + 1 CACAUCCACAUCCACUGCUACUUGCUGUAGCUGCAGCUCCUGCUCC------AGCUCCAGCUGAAGCUAAAGGCAAAGCCAAAGCUUUAUAGCGAAGGCAUCAAGUGCAAGUAUC .............(((((.((((((((.(((((.(((....))).)------)))).))))(((((((...(((...)))..)))))))..((....))...)))))).)))... ( -36.50) >DroYak_CAF1 33911 108 + 1 CACAUCCACAUCCACAGCUAC-------AACUGCAGCUCCAGCUGAAGGAGAAGCUGAAGCUGAAGCUAAAGGCAAAGCCAAAGCUUUAUAGCGAAGGCAUCAAGUGCAAGUAUC ......(((.....(((((.(-------..((.((((....)))).))..).)))))..(((...(((((((((.........))))).))))...))).....)))........ ( -30.40) >consensus CACAUCCACAUCCAUUGCUACU_____________GCUACAGCUAC______AGCUCCAGCUGAAGCUAAAGGCAAAGCCAAAGCUUUAUAGCGAAGGCAUCAAGUGCAAGUAUC ......(((((((.((((((...............(((..((((........))))..)))(((((((...(((...)))..))))))))))))).)).))...)))........ (-21.96 = -22.48 + 0.52)


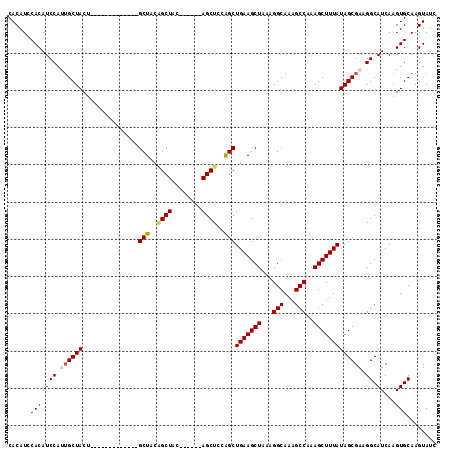
| Location | 3,959,323 – 3,959,419 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.66 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -24.72 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3959323 96 - 23771897 GAUACUUGCACUUGAUGCCUUCGCUAUAAAGCUUUGGCUUUGCCUUUAGCUUCAGCUGGAGCU------GUAGCCGUAGC-------------AGUAGCAAUGGAUGUGGAUGUG ..(((.(.(((......((...(((((...(((.(((((..((.((((((....)))))))).------..))))).)))-------------.)))))...))..))).).))) ( -30.00) >DroSec_CAF1 31799 96 - 1 GAUACUUGCACUUGAUGCCUUCGCUAUAAAGCUUUGGCUUUGCCUUUAGCUUCAGCUGGAGCU------GUAGCUGUAGC-------------AGUAGCAAUGGAUGUGGAUGUG ..(((.(.(((......((...(((((.(((((..(((...)))...)))))..((((.(((.------...))).))))-------------.)))))...))..))).).))) ( -29.00) >DroSim_CAF1 35086 96 - 1 GAUACUUGCACUUGAUGCCUUCGCUAUAAAGCUUUGGCUUUGCCUUUAGCUUCAGCCGGAGCU------GUAGCUGUAGC-------------AGUAGCAAUGGAUGUGGAUGUG .(((.(..((((...(((..(.((((((.((((((((((..((.....))...))))))))))------.....))))))-------------.)..)))..)).))..).))). ( -29.40) >DroEre_CAF1 32696 109 - 1 GAUACUUGCACUUGAUGCCUUCGCUAUAAAGCUUUGGCUUUGCCUUUAGCUUCAGCUGGAGCU------GGAGCAGGAGCUGCAGCUACAGCAAGUAGCAGUGGAUGUGGAUGUG ..(((.(.((((.((.....))(((((.(((((..(((...)))...)))))..((((.((((------(.(((....))).))))).))))..))))))))).).)))...... ( -39.40) >DroYak_CAF1 33911 108 - 1 GAUACUUGCACUUGAUGCCUUCGCUAUAAAGCUUUGGCUUUGCCUUUAGCUUCAGCUUCAGCUUCUCCUUCAGCUGGAGCUGCAGUU-------GUAGCUGUGGAUGUGGAUGUG ..(((.(.(((......((...(((((((((((..(((...)))...)))).(((((((((((........)))))))))))...))-------)))))...))..))).).))) ( -36.80) >consensus GAUACUUGCACUUGAUGCCUUCGCUAUAAAGCUUUGGCUUUGCCUUUAGCUUCAGCUGGAGCU______GUAGCUGUAGC_____________AGUAGCAAUGGAUGUGGAUGUG ..(((.(.(((......((...(((((.(((((..(((...)))...)))))..((((.((((........)))).))))..............)))))...))..))).).))) (-24.72 = -25.28 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:32 2006