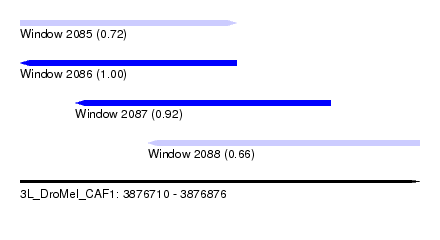
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,876,710 – 3,876,876 |
| Length | 166 |
| Max. P | 0.999056 |

| Location | 3,876,710 – 3,876,800 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 77.70 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3876710 90 + 23771897 GCGCAAGGAGGCGUCGAAGGCGGCAAUGGAACGCUCGCCCCCGCCUAUCCCCGGAGAAGUCCAGGGGCGGAGCGAAGGAGACGCUCCUGC ..(((.((((.((((...((((((........)).))))(((((((..(((.(((....))).)))..)).)))..)).))))))))))) ( -41.10) >DroSec_CAF1 8770 88 + 1 GCGCAAGGAGGCGUCGAAGGCGGCAAUGGAACGCUCGCCCCCGCCCACCCCCGG--UAGUCCAGGGGCGGAGCGAAGGAGACGCUCCUGC ..(((.((((.((((...((((((........)).))))(((((((.((((.((--....)).)))).)).)))..)).))))))))))) ( -44.30) >DroEre_CAF1 5301 88 + 1 GCGCAAGGAGGCGUCGAAGGCGGCAAUGGAACGCUCGCCCCCGCCCACCACCGG--AGGUCCGGGGGCGGAGCGAAGGAGACGCUCCUGC ..(((.((((.((((...((((((........)).))))(((((((.((.((((--....)))).)).)).)))..)).))))))))))) ( -44.00) >DroYak_CAF1 345 88 + 1 GCGCAAGGAGGCGUCGAAGGCGGCAAUGGAACGCUCGCCCCCGCCCACCACUGG--AAAUCCAGGAGCGGAGCGAAGGAGACGCUCCUGC ..(((.((((.((((...((((((........)).))))(((((((.(..((((--....))))..).)).)))..)).))))))))))) ( -40.30) >DroMoj_CAF1 342 71 + 1 GCGUUUGCACCCAUCGAAGACGGCAAUUCGACGCGCGCCCCCGCCCACG------------------AAGAGCGUAGG-AAUGGGCAAAC ((((((((...(......)...)))....)))))..((((...((.(((------------------.....))).))-...)))).... ( -20.70) >consensus GCGCAAGGAGGCGUCGAAGGCGGCAAUGGAACGCUCGCCCCCGCCCACCACCGG__AAGUCCAGGGGCGGAGCGAAGGAGACGCUCCUGC ..(((.((.((((((...((((((........)).))))(((((((.((.((((......)))).)).)).)))..)).))))))))))) (-28.70 = -28.98 + 0.28)

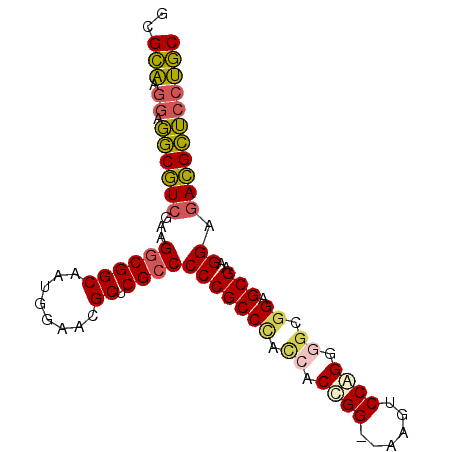
| Location | 3,876,710 – 3,876,800 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 77.70 |
| Mean single sequence MFE | -47.36 |
| Consensus MFE | -41.10 |
| Energy contribution | -39.90 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3876710 90 - 23771897 GCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUUCUCCGGGGAUAGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGACGCCUCCUUGCGC (((((((((((......(((...((((.(((....)))))))...)))(((((((.(((......)))))))))))))).)))).))).. ( -46.70) >DroSec_CAF1 8770 88 - 1 GCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUA--CCGGGGGUGGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGACGCCUCCUUGCGC (((((((((((......((.((((((((((....--))))))))))))(((((((.(((......)))))))))))))).)))).))).. ( -56.30) >DroEre_CAF1 5301 88 - 1 GCAGGAGCGUCUCCUUCGCUCCGCCCCCGGACCU--CCGGUGGUGGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGACGCCUCCUUGCGC (((((((((((......((.(((((.((((....--)))).)))))))(((((((.(((......)))))))))))))).)))).))).. ( -52.40) >DroYak_CAF1 345 88 - 1 GCAGGAGCGUCUCCUUCGCUCCGCUCCUGGAUUU--CCAGUGGUGGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGACGCCUCCUUGCGC (((((((((((......((.((((..((((....--))))..))))))(((((((.(((......)))))))))))))).)))).))).. ( -49.70) >DroMoj_CAF1 342 71 - 1 GUUUGCCCAUU-CCUACGCUCUU------------------CGUGGGCGGGGGCGCGCGUCGAAUUGCCGUCUUCGAUGGGUGCAAACGC ((((((((...-((((((.....------------------))))))((((((((.(((......)))))))))))...)).)))))).. ( -31.70) >consensus GCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUU__CCGGGGGUGGGCGGGGGCGAGCGUUCCAUUGCCGCCUUCGACGCCUCCUUGCGC (((((.(((((......((.(((((.((((......)))).)))))))(((((((.(((......)))))))))))))))...))))).. (-41.10 = -39.90 + -1.20)


| Location | 3,876,733 – 3,876,839 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -49.92 |
| Consensus MFE | -37.73 |
| Energy contribution | -38.57 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3876733 106 - 23771897 GUUUCCGAU-CACGAAACUCAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUUCUCCGGGGAUAGGCGGGGGCGAGCGUUCCAUUG (((((....-...))))).....(((((.(((...))))))))((((((((.((((((((...((((.(((....)))))))...)))))))).)).)))))).... ( -50.40) >DroVir_CAF1 342 87 - 1 GUUCCA-AUGUUAGGAGCUUGAUUGCAGACGCAUUGCGCCGUUGGCUCAUC-CAUACGCUCUU------------------CGUGGGUGGGGGCGUGCGCUACAUUG (((((.-......))))).....(((....)))..((((((((..((((((-(((........------------------.))))))))))))).))))....... ( -28.60) >DroSec_CAF1 8793 104 - 1 GUUUCCGAU-CACGAAACUGAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUA--CCGGGGGUGGGCGGGGGCGAGCGUUCCAUUG (((((....-...))))).....(((((.(((...))))))))((((((((.(((((((.((((((((((....--))))))))))))))))).)).)))))).... ( -60.00) >DroSim_CAF1 9683 104 - 1 GUUUCCGAU-CACGAAACUCAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUU--CCGGGGGUGGGCGGGGGCGAGCGUUCUAUUG (((((....-...))))).....(((((.(((...))))))))((((((((.(((((((.((((((((((....--))))))))))))))))).)).)))))).... ( -58.40) >DroEre_CAF1 5324 104 - 1 GUUCCCGAU-CACGGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCCGGACCU--CCGGUGGUGGGCGGGGGCGAGCGUUCCAUUG (((((....-...))))).....(((.(.(((...)))).)))((((((((.(((((((.(((((.((((....--)))).)))))))))))).)).)))))).... ( -52.40) >DroYak_CAF1 368 104 - 1 GUUCCCGAU-CACGGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCUCCUGGAUUU--CCAGUGGUGGGCGGGGGCGAGCGUUCCAUUG (((((....-...))))).....(((.(.(((...)))).)))((((((((.(((((((.((((..((((....--))))..))))))))))).)).)))))).... ( -49.70) >consensus GUUCCCGAU_CACGAAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUU__CCGGGGGUGGGCGGGGGCGAGCGUUCCAUUG (((((........))))).....(((.(.(((...)))).)))((((((((.(((((((.(((((.((((......)))).)))))))))))).)).)))))).... (-37.73 = -38.57 + 0.84)

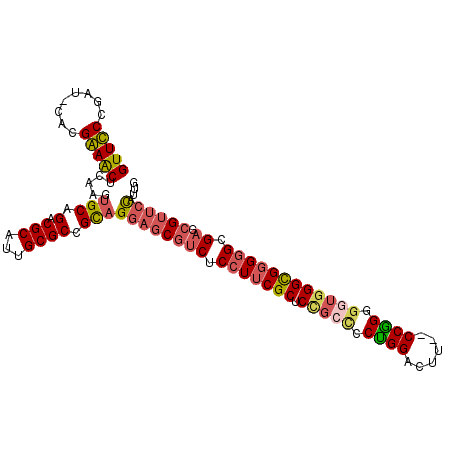
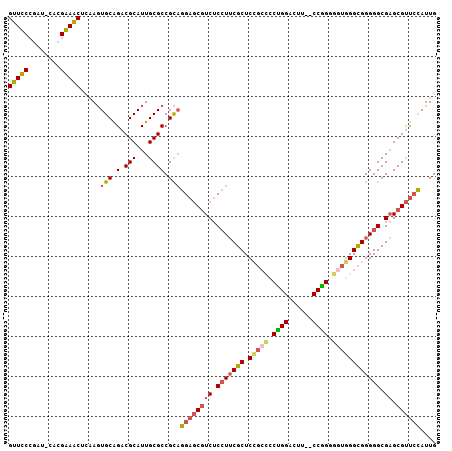
| Location | 3,876,763 – 3,876,876 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -30.09 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3876763 113 - 23771897 AGGAGAAGAGCUUUAAAAAGGGUGCUCGUCCUGGCACGUUUCCGAUCACGAAACUCAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUUCUC ..(((((((((............))))((((.(((..(((((.......))))).....(((((.(((...))))))))((((((.......)))))))))...))))))))) ( -45.20) >DroSec_CAF1 8823 111 - 1 UCGGAAAGAGCUUUAAAAAGGGUGCUCGUCCUGGCACGUUUCCGAUCACGAAACUGAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUA--C .......((((............))))((((.(((..(((((.......))))).....(((((.(((...))))))))((((((.......)))))))))...))))..--. ( -41.60) >DroSim_CAF1 9713 111 - 1 UCGGAAAGAGCUUUAAAAAGGGUGCUCGUCCUGGCACGUUUCCGAUCACGAAACUCAAGUGCGGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUU--C .......((((............))))((((.(((..(((((.......))))).....(((((.(((...))))))))((((((.......)))))))))...))))..--. ( -41.60) >DroEre_CAF1 5354 110 - 1 CCACAUAGAGUUUUAA-AAGGGCGCUCGUCCUGGCACGUUCCCGAUCACGGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCCGGACCU--C ((..............-...(((((.((((...(((((((((.......)))))....)))).))))....)))))((.((((((.......))))))))....))....--. ( -39.00) >DroYak_CAF1 398 110 - 1 GAAUGGAGAGCUUUAA-AAGGGCGCUCGUCCUGGCACGUUCCCGAUCACGGAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCUCCUGGAUUU--C ....((.((((.....-...(((((.((((...(((((((((.......)))))....)))).))))....)))))...((((((.......))))))))))))......--. ( -42.80) >DroAna_CAF1 29618 93 - 1 UUGGCAAAUGUGUUAA-AAGGGUGCUCGUCUUGGCGCGUU--CGAUCGCUGAACAAAAGUGCAGACGCAUUGCGCCGCAGGACCAUCUCAUUUUCU----------------- ((((((....))))))-..(((((...(((((((((((((--((.....)))))...(((((....)))))))))))..)))))))))........----------------- ( -27.40) >consensus UCGGAAAGAGCUUUAA_AAGGGUGCUCGUCCUGGCACGUUUCCGAUCACGAAACUCAAGUGCAGACGCAUUGCGCCGCAGGAGCGUCUCCUUCGCUCCGCCCCUGGACUU__C ....................(((((.(((((..(((((((((.......)))))....)))))))))....)))))((.((((((.......))))))))............. (-30.09 = -30.62 + 0.53)

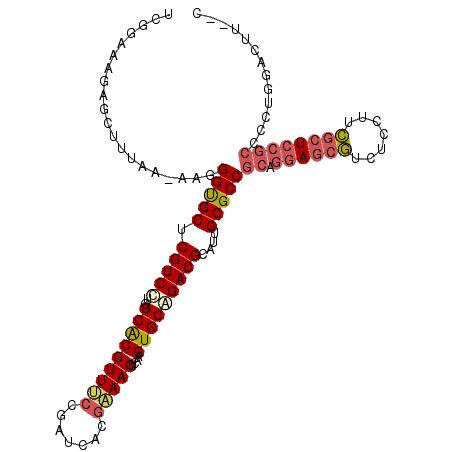

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:07 2006