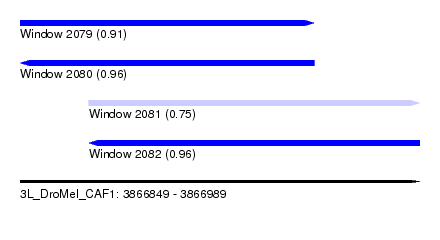
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,866,849 – 3,866,989 |
| Length | 140 |
| Max. P | 0.962315 |

| Location | 3,866,849 – 3,866,952 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -19.39 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3866849 103 + 23771897 AACCGA-UCAUUUUCCAUA---------------UCAUUUUCACAACCGUUUUCACGCAGAUUCACGCCCCCAAAAAAGUCCACUCUAUUAGAGUGGACCGCCGUUAGGUUGC-CAUGAU ......-...........(---------------((((.....(((((........((........))..........((((((((.....))))))))........))))).-.))))) ( -22.00) >DroSec_CAF1 65942 117 + 1 AACCGA-UCAUUUUCCAUAUCCAUAUGUAUAUUAUCAUUUUCACAACCGUUUUCACGCUGAUUCACGCCCACAA-AAGGUCCACUCUAUUAGAGUGGACCGCCGUUAGGUUGC-CAUGAU .....(-((((....((((....))))................(((((........((........))......-..(((((((((.....))))))))).......))))).-.))))) ( -27.00) >DroEre_CAF1 68234 118 + 1 AACCGAAACCUCUUCCACAUGUACGAGUAUAUUCUCAUUUUCACAACCGUUUUCACGCCGAUUCACGCCCACAA-AUGGGCCACUCUAUUAGAGUGGACCGCCGCUAGGUUGC-CGUGAU ...............(((......(((......))).......(((((((....))((.(..((..(((((...-.)))))(((((.....)))))))...).))..))))).-.))).. ( -27.40) >DroYak_CAF1 68383 114 + 1 AACCGA-UCAUCUUCCAUAUAU----GUAUAUUCCCAUAUUCACAAUCGUUUUCACGCUGAUUCACGCCCACAA-AGAGUCCACUCUAUUAGAGUGGACCACCGUUAGGCUGAAGGUGAU .....(-((((((((...((((----(........)))))..........................(((.....-...((((((((.....))))))))........))).))))))))) ( -28.79) >consensus AACCGA_UCAUCUUCCAUAU_U____GUAUAUUCUCAUUUUCACAACCGUUUUCACGCUGAUUCACGCCCACAA_AAAGUCCACUCUAUUAGAGUGGACCGCCGUUAGGUUGC_CAUGAU ..................................((((.....(((((........((........)).........(((((((((.....))))))))).......)))))...)))). (-19.39 = -20.20 + 0.81)


| Location | 3,866,849 – 3,866,952 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -25.58 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3866849 103 - 23771897 AUCAUG-GCAACCUAACGGCGGUCCACUCUAAUAGAGUGGACUUUUUUGGGGGCGUGAAUCUGCGUGAAAACGGUUGUGAAAAUGA---------------UAUGGAAAAUGA-UCGGUU .(((((-.(..(((((..(.(((((((((.....))))))))))..)))))).))))).(((.(((....)))((..(....)..)---------------)..)))......-...... ( -31.50) >DroSec_CAF1 65942 117 - 1 AUCAUG-GCAACCUAACGGCGGUCCACUCUAAUAGAGUGGACCUU-UUGUGGGCGUGAAUCAGCGUGAAAACGGUUGUGAAAAUGAUAAUAUACAUAUGGAUAUGGAAAAUGA-UCGGUU .(((..-((..((((.....(((((((((.....)))))))))..-...))))..........(((....)))))..)))...((((......((((....)))).......)-)))... ( -34.82) >DroEre_CAF1 68234 118 - 1 AUCACG-GCAACCUAGCGGCGGUCCACUCUAAUAGAGUGGCCCAU-UUGUGGGCGUGAAUCGGCGUGAAAACGGUUGUGAAAAUGAGAAUAUACUCGUACAUGUGGAAGAGGUUUCGGUU .(((((-((((((..(((.((((.(((((.....)))))(((((.-...)))))....)))).)))......))))))....(((((......)))))...))))).............. ( -37.90) >DroYak_CAF1 68383 114 - 1 AUCACCUUCAGCCUAACGGUGGUCCACUCUAAUAGAGUGGACUCU-UUGUGGGCGUGAAUCAGCGUGAAAACGAUUGUGAAUAUGGGAAUAUAC----AUAUAUGGAAGAUGA-UCGGUU ((((.((((.(((((.(((.(((((((((.....)))))))))..-)))))))).....(((.(((....)))....)))(((((........)----))))...)))).)))-)..... ( -37.30) >consensus AUCACG_GCAACCUAACGGCGGUCCACUCUAAUAGAGUGGACCUU_UUGUGGGCGUGAAUCAGCGUGAAAACGGUUGUGAAAAUGAGAAUAUAC____A_AUAUGGAAAAUGA_UCGGUU .((((..((((((.......(((((((((.....))))))))).........((((......))))......))))))....)))).................................. (-25.58 = -26.20 + 0.62)


| Location | 3,866,873 – 3,866,989 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.48 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3866873 116 + 23771897 UCACAACCGUUUUCACGCAGAUUCACGCCCCCAAAAAAGUCCACUCUAUUAGAGUGGACCGCCGUUAGGUUGC-CAUGAUCUGAUCUCACUCAUUUUCCCAGGCCCAAAGCUAUAAA .................((((((...((..........((((((((.....)))))))).(((....))).))-...))))))..................(((.....)))..... ( -24.70) >DroSec_CAF1 65981 115 + 1 UCACAACCGUUUUCACGCUGAUUCACGCCCACAA-AAGGUCCACUCUAUUAGAGUGGACCGCCGUUAGGUUGC-CAUGAUCUGAUCUCACUCAUUUUCCCAGGCCCAAAGCUGUAAA ................(((.......(((.....-..(((((((((.....)))))))))(..((((((((..-...))))))))..).............)))....)))...... ( -29.60) >DroEre_CAF1 68274 113 + 1 UCACAACCGUUUUCACGCCGAUUCACGCCCACAA-AUGGGCCACUCUAUUAGAGUGGACCGCCGCUAGGUUGC-CGUGAUCGG--CUCACCCAUUUUCCCAGGCCCAAAGCCAUAUA ................((((((.((((((((...-.)))))(((((.....)))))((((.......))))..-.))))))))--)...............(((.....)))..... ( -35.90) >DroYak_CAF1 68418 114 + 1 UCACAAUCGUUUUCACGCUGAUUCACGCCCACAA-AGAGUCCACUCUAUUAGAGUGGACCACCGUUAGGCUGAAGGUGAUCUG--CUCACCCAUUUUCCCAGGCCCAAAGCUAUAUA ........(((((...(((..((((.(((.....-...((((((((.....))))))))........)))))))(((((....--.)))))..........)))..)))))...... ( -31.29) >consensus UCACAACCGUUUUCACGCUGAUUCACGCCCACAA_AAAGUCCACUCUAUUAGAGUGGACCGCCGUUAGGUUGC_CAUGAUCUG__CUCACCCAUUUUCCCAGGCCCAAAGCUAUAAA ...(((((........((........)).........(((((((((.....))))))))).......))))).............................(((.....)))..... (-21.23 = -21.85 + 0.62)


| Location | 3,866,873 – 3,866,989 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 89.48 |
| Mean single sequence MFE | -40.55 |
| Consensus MFE | -33.85 |
| Energy contribution | -33.47 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.962002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3866873 116 - 23771897 UUUAUAGCUUUGGGCCUGGGAAAAUGAGUGAGAUCAGAUCAUG-GCAACCUAACGGCGGUCCACUCUAAUAGAGUGGACUUUUUUGGGGGCGUGAAUCUGCGUGAAAACGGUUGUGA .(((((((((((.(((...(...((((.((....))..)))).-.)..(((((..(.(((((((((.....))))))))))..)))))))).))))....(((....)))))))))) ( -36.70) >DroSec_CAF1 65981 115 - 1 UUUACAGCUUUGGGCCUGGGAAAAUGAGUGAGAUCAGAUCAUG-GCAACCUAACGGCGGUCCACUCUAAUAGAGUGGACCUU-UUGUGGGCGUGAAUCAGCGUGAAAACGGUUGUGA .(((((((((((.(((((.((((.((.((((.......)))).-.)).((....)).(((((((((.....)))))))))))-)).))))).))))....(((....)))))))))) ( -42.10) >DroEre_CAF1 68274 113 - 1 UAUAUGGCUUUGGGCCUGGGAAAAUGGGUGAG--CCGAUCACG-GCAACCUAGCGGCGGUCCACUCUAAUAGAGUGGCCCAU-UUGUGGGCGUGAAUCGGCGUGAAAACGGUUGUGA ..((..(((....(((((......)))))..(--(((((((((-((..((....))..)))(((((.....)))))(((((.-...)))))))).))))))((....)))))..)). ( -43.40) >DroYak_CAF1 68418 114 - 1 UAUAUAGCUUUGGGCCUGGGAAAAUGGGUGAG--CAGAUCACCUUCAGCCUAACGGUGGUCCACUCUAAUAGAGUGGACUCU-UUGUGGGCGUGAAUCAGCGUGAAAACGAUUGUGA ........((((.(((((((.....((((((.--....))))))....))).((((.(((((((((.....)))))))))..-)))))))).)))).((((((....))).)))... ( -40.00) >consensus UAUAUAGCUUUGGGCCUGGGAAAAUGAGUGAG__CAGAUCACG_GCAACCUAACGGCGGUCCACUCUAAUAGAGUGGACCUU_UUGUGGGCGUGAAUCAGCGUGAAAACGGUUGUGA ..((((((((((.(((((((.......((((.......))))......)))......(((((((((.....))))))))).......)))).))))....(((....))))))))). (-33.85 = -33.47 + -0.38)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:02 2006