| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 419,853 – 419,945 |
| Length | 92 |
| Max. P | 0.810398 |

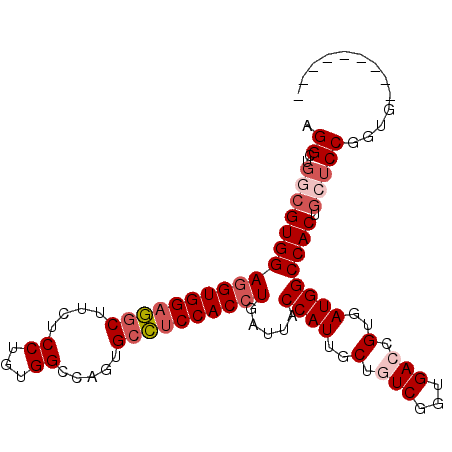
| Location | 419,853 – 419,945 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -40.43 |
| Consensus MFE | -32.41 |
| Energy contribution | -33.63 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 419853 92 + 23771897 AGGCUGGCGUGGAGGUGGAGGCUUCUCCUGUGGCUAGUGCCUCCACCUGAUUACCAUUGCUGUCGGUGACCGUGAUGGCCACUGCUCCGGUA--------- ..(((((.(..(((((((((((....((...)).....)))))))))).....((((..(.(((...))).)..))))...)..).))))).--------- ( -41.30) >DroSec_CAF1 1205 92 + 1 AGGCUGGCGUGGAGGUGGAGGCUUCUCCUGUGGCUAGUGCCUCCACCUGAUUACCAUUGCUAUCGGUGACCGUGAUGGCCACUGCUCCGGAG--------- .((..(((((((((((((((((....((...)).....)))))))))).....((((..(....(....).)..)))))))).)))))....--------- ( -38.30) >DroSim_CAF1 1205 92 + 1 AGGCUGGCGUGGAGGUGGAGGCUUCUCCUGUGGCUAGUGCCUCCACCUGAUUACCAUUGCUAUCGGUGACCGUGAUGGCCACUGCUCCGGAG--------- .((..(((((((((((((((((....((...)).....)))))))))).....((((..(....(....).)..)))))))).)))))....--------- ( -38.30) >DroEre_CAF1 1207 92 + 1 AGGCUGUCGUGGAGGUGGCGGCUUCUCCUGUGGCCAGAGCCUCCACCUGAUUGCCAUUGCUGUCGGUGACCGUGAUGGCCACUGCUCCGCUG--------- .((((((((((((((((((.((.......)).)))...))))))))(((((.((....)).))))).......)))))))............--------- ( -38.60) >DroYak_CAF1 1214 92 + 1 AGGCUGGCGUGGAGGUGGAGGCUUCUCCAGUGGCCAGAGCCUCCACCUGAUUGCCAUUGCUGUCGGUGACCGUGAUGGCCACAGCUCCGUUG--------- .((..((((((.((((((((((((..((...))...))))))))))))....(((((..(.(((...))).)..)))))))).)))))....--------- ( -43.20) >DroPer_CAF1 49234 101 + 1 AGGCACCAGUGGAAGUGGAAGCCUCUCCGGUGGCCAACGAUUCCACCUGGUUGCCAUUGCUGUCGGUGACGGUGAUGGCCACCGAUCCGUUGAGCUCUUUC .(((((((((((((.(((..(((.....)))..)))....))))).)))).))))...(((..(((...(((((.....)))))..)))...)))...... ( -42.90) >consensus AGGCUGGCGUGGAGGUGGAGGCUUCUCCUGUGGCCAGUGCCUCCACCUGAUUACCAUUGCUGUCGGUGACCGUGAUGGCCACUGCUCCGGUG_________ .((..(((((((((((((((((....((...)).....)))))))))).....((((..(.(((...))).)..)))))))).)))))............. (-32.41 = -33.63 + 1.22)

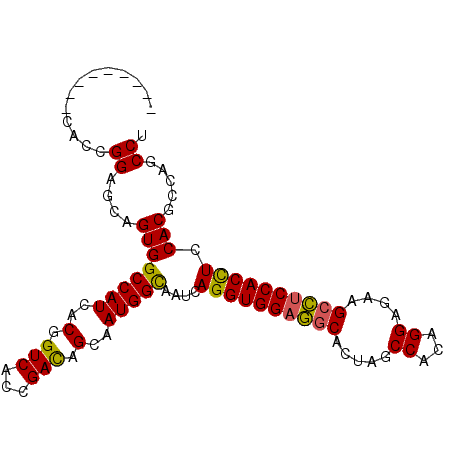
| Location | 419,853 – 419,945 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -30.17 |
| Energy contribution | -29.62 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 419853 92 - 23771897 ---------UACCGGAGCAGUGGCCAUCACGGUCACCGACAGCAAUGGUAAUCAGGUGGAGGCACUAGCCACAGGAGAAGCCUCCACCUCCACGCCAGCCU ---------....((.((.((((((.....)))))).........((((....((((((((((.....((...))....))))))))))....)))))))) ( -35.40) >DroSec_CAF1 1205 92 - 1 ---------CUCCGGAGCAGUGGCCAUCACGGUCACCGAUAGCAAUGGUAAUCAGGUGGAGGCACUAGCCACAGGAGAAGCCUCCACCUCCACGCCAGCCU ---------....((.((.((((((.....)))))).........((((....((((((((((.....((...))....))))))))))....)))))))) ( -35.40) >DroSim_CAF1 1205 92 - 1 ---------CUCCGGAGCAGUGGCCAUCACGGUCACCGAUAGCAAUGGUAAUCAGGUGGAGGCACUAGCCACAGGAGAAGCCUCCACCUCCACGCCAGCCU ---------....((.((.((((((.....)))))).........((((....((((((((((.....((...))....))))))))))....)))))))) ( -35.40) >DroEre_CAF1 1207 92 - 1 ---------CAGCGGAGCAGUGGCCAUCACGGUCACCGACAGCAAUGGCAAUCAGGUGGAGGCUCUGGCCACAGGAGAAGCCGCCACCUCCACGACAGCCU ---------..((....(.((((((.....)))))).)...))...(((..((((((((.(((((((....)))....)))).))))))....))..))). ( -35.10) >DroYak_CAF1 1214 92 - 1 ---------CAACGGAGCUGUGGCCAUCACGGUCACCGACAGCAAUGGCAAUCAGGUGGAGGCUCUGGCCACUGGAGAAGCCUCCACCUCCACGCCAGCCU ---------....((.(((((((((.....))))....)))))..((((....(((((((((((....((...))...)))))))))))....)))).)). ( -40.30) >DroPer_CAF1 49234 101 - 1 GAAAGAGCUCAACGGAUCGGUGGCCAUCACCGUCACCGACAGCAAUGGCAACCAGGUGGAAUCGUUGGCCACCGGAGAGGCUUCCACUUCCACUGGUGCCU ......(((...(((..(((((.....)))))...)))..)))...((((.((((.(((((..(..((((........))))..)..))))))))))))). ( -39.70) >consensus _________CACCGGAGCAGUGGCCAUCACGGUCACCGACAGCAAUGGCAAUCAGGUGGAGGCACUAGCCACAGGAGAAGCCUCCACCUCCACGCCAGCCU .............((....((((((((..(.(((...))).)..)))))....((((((((((.....((...))....)))))))))).))).....)). (-30.17 = -29.62 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:29 2006