| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,853,890 – 3,853,990 |
| Length | 100 |
| Max. P | 0.949052 |

| Location | 3,853,890 – 3,853,990 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -17.63 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
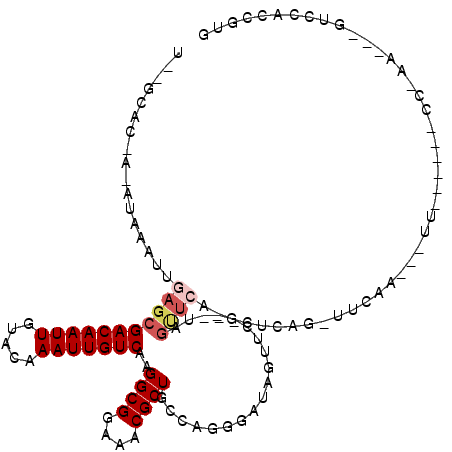
>3L_DroMel_CAF1 3853890 100 + 23771897 U--GUAC-AUAUAAAUUGAGCGACAAUUGUACAAAUUGUCAAGGCGGAAACGCUGGCAGGGAUAAAA----UAGUUCGAGCUCGGAUUCCA---UU------CC-AU---GUCCACUGUG .--((.(-(.......)).))(((((((.....)))))))..((((....)))).(((((((((...----.(((....))).(((.....---.)------))-.)---)))).)))). ( -29.70) >DroVir_CAF1 65648 87 + 1 U--CCAC-A-AUAAAUUGAGCGACAAUUGUACAAAUUGUCAAGGCGGAAACGCUGCCAGGGAUAGUUC---------------AG-UUCAG---UUCG-------GA---GUCCAGCGUG .--.(((-.-...(((((((((((((((.....)))))))..((((....))))..........))))---------------))-))..(---((.(-------(.---..)))))))) ( -26.20) >DroPse_CAF1 60644 105 + 1 UUCAUACUAUAUAAAUUGAGCGACAAUUGUACAAAUUGUCAAGGCGGAAACGCUGCCAGGGAUAGUUC---GAGCUCCAGCUCGGAUUCCA---CU------CCGAU---GUCCACUGUG ...................(((((((((.....)))))))..((((....))))))((((((((((((---((((....))))))))....---..------....)---)))).))).. ( -34.50) >DroGri_CAF1 59282 111 + 1 U--CCAC-A-AUAAAUUGAGCGACAAUUGUACAAAUUGUCAAGGCGGAAACGCUGCCAGGGAUAGUUCCAACAGUUCA-AC--AG-UUCAGUAGUUCAGCGAGC-GAGUUGUCCAGCGUG .--.(((-.-.....(((((((((((((.....)))))))..((((....)))).....(((....)))....)))))-).--..-....((.(..((((....-..))))..).))))) ( -32.20) >DroEre_CAF1 54125 100 + 1 U--GCAC-AUAUAAAUUGAGCGACAAUUGUACAAAUUGUCAAGGCGGAAACGCUGGCAGGGAUAAAA----UAGUUGCAGCUCGGAUUCCA---UU------CC-AU---GUCCACUGUG .--((.(-(.......)).))(((((((.....)))))))..((((....)))).(((((((((...----.(((....))).(((.....---.)------))-.)---)))).)))). ( -31.30) >DroMoj_CAF1 66303 103 + 1 U--GCAC-A-AUAAAUUGAGCGACAAUUGUACAAAUUGUCAAGGCGGAAACGCUGCCAGGGAUAGUUC-GGUAGCUCA-ACUCAG-UUCAA---CUCG---GCC-GA---GUCCAGCGUG .--.(((-.-....((((((((((((((.....)))))))..((((....))))..........))))-))).(((..-((((.(-((...---...)---)).-))---))..)))))) ( -33.20) >consensus U__GCAC_A_AUAAAUUGAGCGACAAUUGUACAAAUUGUCAAGGCGGAAACGCUGCCAGGGAUAGUUC___UAGUUCA_GCUCAG_UUCAA___UU______CC_AA___GUCCACCGUG .................(((((((((((.....)))))))..((((....))))...................))))........................................... (-17.63 = -18.30 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:50 2006