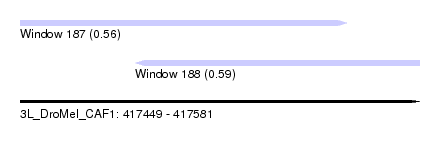
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 417,449 – 417,581 |
| Length | 132 |
| Max. P | 0.591765 |

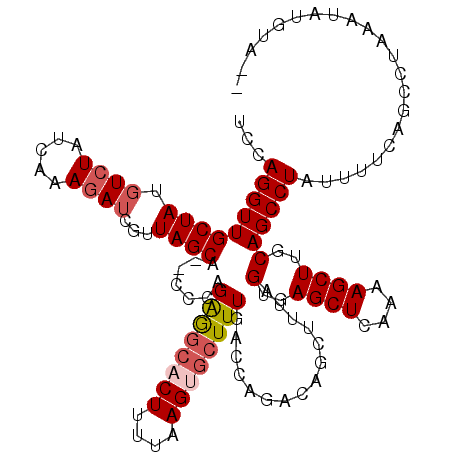
| Location | 417,449 – 417,557 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -19.25 |
| Energy contribution | -20.50 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 417449 108 + 23771897 UCCAGGUUGCUAUGUCUAUAAAAGAUCUUUAGCA--CCCGGAGGCACUUAUAAGCGCUUUGACCAUACAUCUUUAGUGAGCUCAAAAGCUUCCAGCCUAUUUUCAGCCU--------A-- (((.(((.((((.((((.....))))...)))))--)).)))(((......((((..(((((.(.(((.......))).).))))).))))...)))............--------.-- ( -25.00) >DroSec_CAF1 64390 116 + 1 UCCAGGUUGCUAUGUCUUUCAAAGAUCGUUAGCA--CCCAGAGGCACUUUUAAGUGCUUUGACCAAACAGCUUUAGUGAGCUCAAAAGCUUGCAGCCUAUUUUUAGCCUAAAUAUGUA-- ....((.(((((.((((.....))))...)))))--.)).((((((((....))))))))......(((......(..((((....))))..)((.(((....))).)).....))).-- ( -29.10) >DroSim_CAF1 66536 116 + 1 UCCAGGUUGCUAUGUCUUUCAAAGAUCGUUAGCA--CCCAGGGGCACUUUUAAGUGCUUUGACCAGACAGCUUUAGUGAGCUCAAAAGCUUGCAGCCUAUUUUCAGCCUAAAUAUGUA-- ...((((((...(((((.((((((..........--((....))((((....))))))))))..)))))(((...(..((((....))))..)))).......)))))).........-- ( -29.90) >DroYak_CAF1 68861 110 + 1 UCCAGGUUGCUACUUAUAUGAAAGAUUAUUAGCUUCCCCAGAAGCA----------GUUUGUUCCGACAGCUUUCGUGAGCUCAAAAGCUUGCAGCCUAUUUUCAGCCAAAAUAUGUGUA ...(((((((.......(((((((.......(((((....))))).----------(((......)))..)))))))(((((....))))))))))))...................... ( -27.40) >consensus UCCAGGUUGCUAUGUCUAUCAAAGAUCGUUAGCA__CCCAGAGGCACUUUUAAGUGCUUUGACCAGACAGCUUUAGUGAGCUCAAAAGCUUGCAGCCUAUUUUCAGCCUAAAUAUGUA__ ...(((((((((.((((.....))))...)))).......((((((((....))))))))...............(..((((....))))..))))))...................... (-19.25 = -20.50 + 1.25)

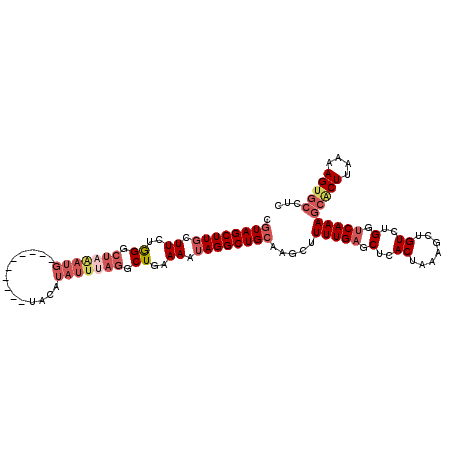
| Location | 417,487 – 417,581 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -15.25 |
| Energy contribution | -18.44 |
| Covariance contribution | 3.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 417487 94 - 23771897 CGUAGCUUGCUUCUAGGAUAAAUG----------U--------AGGCUGAAAAUAGGCUGGAAGCUUUUGAGCUCACUAAAGAUGUAUGGUCAAAGCGCUUAUAAGUGCCUC ..((((((((.............)----------)--------)))))).....((((...((((((((((.(..((.......))..).)))))).))))......)))). ( -23.92) >DroSec_CAF1 64428 102 - 1 CGUAGCUUUCUUCUGGGCUAGAUG----------UACAUAUUUAGGCUAAAAAUAGGCUGCAAGCUUUUGAGCUCACUAAAGCUGUUUGGUCAAAGCACUUAAAAGUGCCUC .(((((((..((.(((.(((((((----------....))))))).))).))..))))))).....(((((.(..((.......))..).)))))(((((....)))))... ( -28.80) >DroSim_CAF1 66574 102 - 1 CGUAGCUUGCUUCUGGGCUAGAUG----------UACAUAUUUAGGCUGAAAAUAGGCUGCAAGCUUUUGAGCUCACUAAAGCUGUCUGGUCAAAGCACUUAAAAGUGCCCC .((((((((.((..((.(((((((----------....))))))).))..)).)))))))).....(((((.(..((.......))..).)))))(((((....)))))... ( -29.80) >DroYak_CAF1 68901 102 - 1 UGUAGCUUGCUUCUGGGCUUAAUGCUUAUUUUUACACAUAUUUUGGCUGAAAAUAGGCUGCAAGCUUUUGAGCUCACGAAAGCUGUCGGAACAAAC----------UGCUUC .((((.(((.((((((((.....(((((((((((..((.....))..))))))))))).)).((((((((......)))))))).))))))))).)----------)))... ( -32.40) >consensus CGUAGCUUGCUUCUGGGCUAAAUG__________UACAUAUUUAGGCUGAAAAUAGGCUGCAAGCUUUUGAGCUCACUAAAGCUGUCUGGUCAAAGCACUUAAAAGUGCCUC .((((((((.((..((.(((((((..............))))))).))..)).)))))))).....(((((.(..((.......))..).)))))(((((....)))))... (-15.25 = -18.44 + 3.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:27 2006