| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
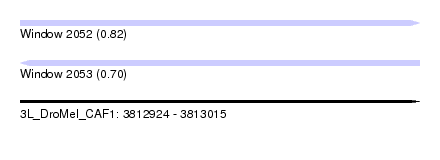
| Location | 3,812,924 – 3,813,015 |
| Length | 91 |
| Max. P | 0.823953 |

| Location | 3,812,924 – 3,813,015 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.44 |
| Mean single sequence MFE | -14.48 |
| Consensus MFE | -11.42 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
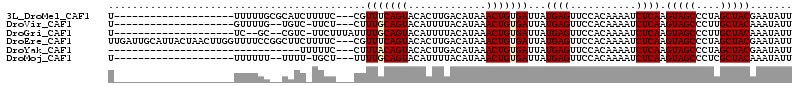
>3L_DroMel_CAF1 3812924 91 + 23771897 U--------------------UUUUUGCGCAUCUUUUC---CGUUUCAGUACACUUGACAUAAACUGUGAUUAUGAGUUCCACAAAAUCUCAAGUAGCCCUAGCUACGAAUAUU .--------------------.(((((.(...(((...---.(((.((((.............)))).)))...)))...).)))))......(((((....)))))....... ( -12.22) >DroVir_CAF1 14360 88 + 1 U--------------------GUUUUG--UGUC-UUCU---CUUUGCAGUACAUUUUACAUAAACUGUGAUUAUGAGUUCCACAAAAUCUCAAGUAGCCCUUGCUACAAAUAUU .--------------------((((((--((..-..((---(.(..((((.............))))..)....)))...)))))))).....(((((....)))))....... ( -20.32) >DroGri_CAF1 13050 89 + 1 U--------------------UC--GC--CGUC-UUCUUUAUUUUGCAGUACAUUUUACAUAAACUGUGAUUAUGAGUUCCACAAAAUCUCAAGUAGCCCUUGCUACAAAUAUU .--------------------..--..--.((.-..(((....(..((((.............))))..)....)))....))..........(((((....)))))....... ( -12.72) >DroEre_CAF1 13149 111 + 1 UUGAUUGCAUUACUAACUUGGUUUUCCGGCUUCUUUUC---CGUUUCAGUACACUUGACAUAAACUGUGAUUAUGAGUUCCACAAAAUCUCAAGUAGCCCUAGCUACGAAUAUU .((((..((........((((....)))).........---.(((..((....)).)))......))..))))((((...........)))).(((((....)))))....... ( -15.60) >DroYak_CAF1 13252 79 + 1 --------------------------------UUUUUC---CUUUACAGUACACUUGACAUAAACUGUGAUUAUGAGUUCCACAAAAUCUCAAGUAGCCCUAGCUACGAAUAUU --------------------------------......---..(((((((.............)))))))...((((...........)))).(((((....)))))....... ( -13.32) >DroMoj_CAF1 14618 88 + 1 U--------------------UUUUUU--UUUU-UGCU---UUUUGCAGUACAUUUUACAUAAACUGUGAUUAUGAGUUCCACAAAAUCUCAAGUAGCCCUCGCUACAAAUAUU .--------------------......--...(-(((.---....))))......(((((.....)))))...((((...........)))).(((((....)))))....... ( -12.70) >consensus U____________________UUUUUC__CGUC_UUCC___CUUUGCAGUACACUUGACAUAAACUGUGAUUAUGAGUUCCACAAAAUCUCAAGUAGCCCUAGCUACAAAUAUU ...........................................(((((((.............)))))))...((((...........)))).(((((....)))))....... (-11.42 = -11.67 + 0.25)
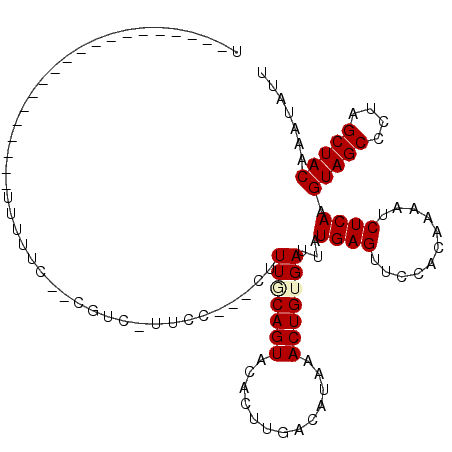

| Location | 3,812,924 – 3,813,015 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.44 |
| Mean single sequence MFE | -17.23 |
| Consensus MFE | -12.75 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3812924 91 - 23771897 AAUAUUCGUAGCUAGGGCUACUUGAGAUUUUGUGGAACUCAUAAUCACAGUUUAUGUCAAGUGUACUGAAACG---GAAAAGAUGCGCAAAAA--------------------A ....(((((..(.((..(.((((((.((.((((((.........))))))...)).)))))))..)))..)))---))...............--------------------. ( -18.30) >DroVir_CAF1 14360 88 - 1 AAUAUUUGUAGCAAGGGCUACUUGAGAUUUUGUGGAACUCAUAAUCACAGUUUAUGUAAAAUGUACUGCAAAG---AGAA-GACA--CAAAAC--------------------A .......(((((....)))))......(((((((...(((.......((((.(((.....))).))))....)---))..-..))--))))).--------------------. ( -16.80) >DroGri_CAF1 13050 89 - 1 AAUAUUUGUAGCAAGGGCUACUUGAGAUUUUGUGGAACUCAUAAUCACAGUUUAUGUAAAAUGUACUGCAAAAUAAAGAA-GACG--GC--GA--------------------A .......(((((....))))).....((((((..(.((.(((((.......)))))......)).)..))))))......-....--..--..--------------------. ( -15.30) >DroEre_CAF1 13149 111 - 1 AAUAUUCGUAGCUAGGGCUACUUGAGAUUUUGUGGAACUCAUAAUCACAGUUUAUGUCAAGUGUACUGAAACG---GAAAAGAAGCCGGAAAACCAAGUUAGUAAUGCAAUCAA ..((((..(((((.((((.((((((.((.((((((.........))))))...)).)))))))).......((---(........))).....)).)))))..))))....... ( -21.80) >DroYak_CAF1 13252 79 - 1 AAUAUUCGUAGCUAGGGCUACUUGAGAUUUUGUGGAACUCAUAAUCACAGUUUAUGUCAAGUGUACUGUAAAG---GAAAAA-------------------------------- ....(((...((.((..(.((((((.((.((((((.........))))))...)).)))))))..))))...)---))....-------------------------------- ( -15.30) >DroMoj_CAF1 14618 88 - 1 AAUAUUUGUAGCGAGGGCUACUUGAGAUUUUGUGGAACUCAUAAUCACAGUUUAUGUAAAAUGUACUGCAAAA---AGCA-AAAA--AAAAAA--------------------A ....(((((((.(((..((((..........))))..)))......(((.....)))........))))))).---....-....--......--------------------. ( -15.90) >consensus AAUAUUCGUAGCUAGGGCUACUUGAGAUUUUGUGGAACUCAUAAUCACAGUUUAUGUAAAAUGUACUGCAAAG___AAAA_GAAG__CAAAAA____________________A .......(((((....))))).((((.(((....)))))))......((((.(((.....))).)))).............................................. (-12.75 = -12.50 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:34 2006