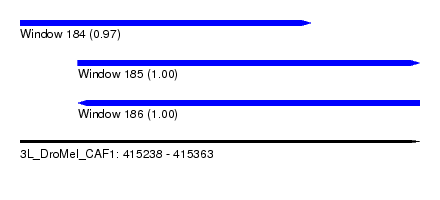
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 415,238 – 415,363 |
| Length | 125 |
| Max. P | 0.999046 |

| Location | 415,238 – 415,329 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 73.81 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.85 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
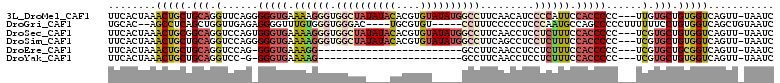
>3L_DroMel_CAF1 415238 91 + 23771897 UGGCUUUCCUCACAUCACUUCACUAAACUGCUGCAGGUUCAGGGGGUGAAAAGGGUGGCUAUAUACACGUGUAUAUGGCCUUCAACAUCCC ......((((....(((((((.((.((((......)))).)))))))))..)))).((((((((((....))))))))))........... ( -28.30) >DroGri_CAF1 73949 80 + 1 UCAGUUUAUUCACUUGGCUGCAC--AGCCUCAGCUGGUUGAGAGGGGUUUGUGGGUGGGAC----UGCGUGU-----CCUUUCCCCCUCCC .........(((.(..((((...--.....))))..).)))((((((......((.(((((----.....))-----))).)))))))).. ( -26.50) >DroSec_CAF1 62157 91 + 1 UGGCUUUCCUCACUUCACUUCACUAAACUGCGGCAGGUCCAGUGGGUGAAAAGGGUGGCUAUAUACACGUGUAUAUGGCCUUCAACCUCCU .((...((((...((((((.((((..(((......)))..)))))))))).)))).((((((((((....)))))))))).....)).... ( -29.30) >DroSim_CAF1 64292 91 + 1 UGGCUUUCCUCACUUCACUUCACUAAACUGCUGCAGGUCCAGGGGGUGAAAAGGGUGGCUAUAUACACGUGUAUAUGGCCUUCAGCCUCCU .((((.((((...((((((((.((..(((......)))..)))))))))).)))).((((((((((....))))))))))...)))).... ( -33.30) >consensus UGGCUUUCCUCACUUCACUUCACUAAACUGCUGCAGGUCCAGGGGGUGAAAAGGGUGGCUAUAUACACGUGUAUAUGGCCUUCAACCUCCC ........(((...(((((((.((.((((......)))).)))))))))...))).((((((((((....))))))))))........... (-17.60 = -18.85 + 1.25)

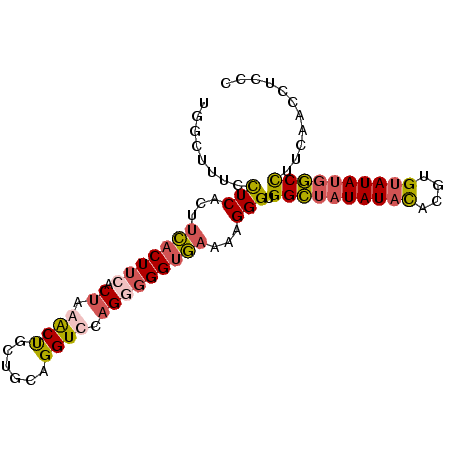
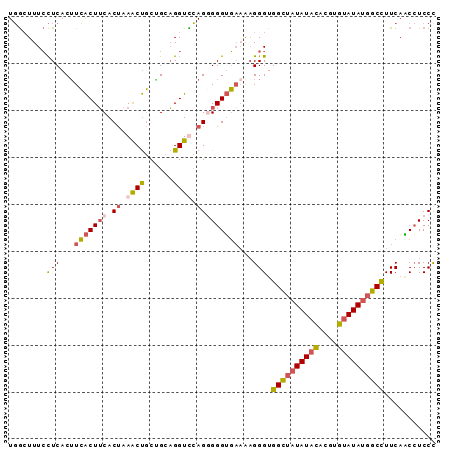
| Location | 415,256 – 415,363 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -19.81 |
| Energy contribution | -22.25 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.40 |
| Mean z-score | -4.27 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
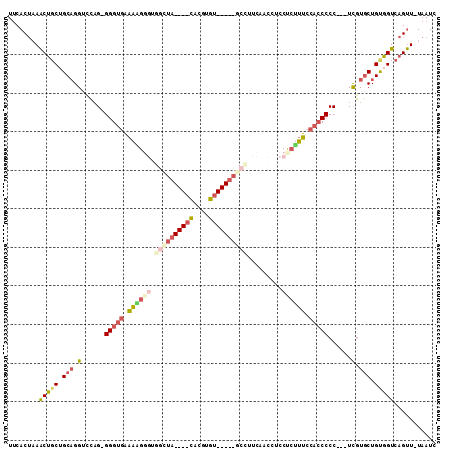
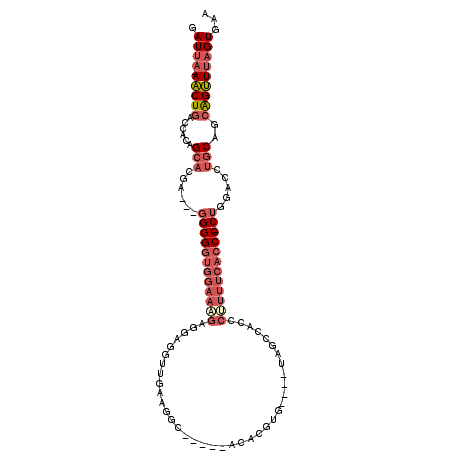
>3L_DroMel_CAF1 415256 107 + 23771897 UUCACUAAACUGCUGCAGGUUCAGGGGGUGAAAAGGGUGGCUAUAUACACGUGUAUAUGGCCUUCAACAUCCCCAUUCCACCCCC---UUGUGCUGUGGUCAGUU-UAAUC .....((((((((..(((....((((((((.((.(((.((((((((((....)))))))))).........))).)).)))))))---)....)))..).)))))-))... ( -48.20) >DroGri_CAF1 73967 100 + 1 UGCAC--AGCCUCAGCUGGUUGAGAGGGGUUUGUGGGUGGGAC----UGCGUGU-----CCUUUCCCCCUCCCAAUGCCAGCCCCCUUUUUUCCUGUGGUCAGCUGUAAUC ...((--(((....((.((..((((((((.(((((((.(((((----.....))-----)))........))))....))).))))))))..)).)).....))))).... ( -36.00) >DroSec_CAF1 62175 107 + 1 UUCACUAAACUGCGGCAGGUCCAGUGGGUGAAAAGGGUGGCUAUAUACACGUGUAUAUGGCCUUCAACCUCCUCUUUCCACCCCC---UCGUGCUGUGGUCAGUU-UAAUC .....((((((((.((((..(.((.(((((.((((((.((((((((((....)))))))))).........)))))).))))).)---).)..)))).).)))))-))... ( -42.10) >DroSim_CAF1 64310 107 + 1 UUCACUAAACUGCUGCAGGUCCAGGGGGUGAAAAGGGUGGCUAUAUACACGUGUAUAUGGCCUUCAGCCUCCUCUUUCCACCCCC---UCGUGCUGUGGUCAGUU-UAAUC .....((((((((..(((..(.((((((((.((((((.((((((((((....)))))))))).........)))))).)))))))---).)..)))..).)))))-))... ( -50.10) >DroEre_CAF1 66427 82 + 1 UUCACUAAACUGCUGCAGGUCCAG-GGGUGAAAGG------------------------GCCUUCAACCUCCUCUUUCCACCCCC---UCGUGCUGCGGUCAGUU-UAAUC .....(((((((((((((.....(-((((((((((------------------------(...........)))))).)))))).---.....)))))).)))))-))... ( -31.90) >DroYak_CAF1 66602 81 + 1 UUCACUAAACUGCUGCAGGUCC-G-GGGUGAAAAG------------------------GCCUUCAACCUCCUCUUUCCACCCCC---UCGUGCUGUGGUCAGUU-UAAUC .....((((((((..(((....-(-(((((.((((------------------------(............))))).)))))).---.....)))..).)))))-))... ( -26.60) >consensus UUCACUAAACUGCUGCAGGUCCAG_GGGUGAAAAGGGUGGCUA____CACGUGU_____GCCUUCAACCUCCUCUUUCCACCCCC___UCGUGCUGUGGUCAGUU_UAAUC ........(((((.(((.(......(((((.((((((.((((((((((....)))))))))).........)))))).)))))......).))).)))))........... (-19.81 = -22.25 + 2.44)

| Location | 415,256 – 415,363 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -11.66 |
| Energy contribution | -13.58 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.30 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
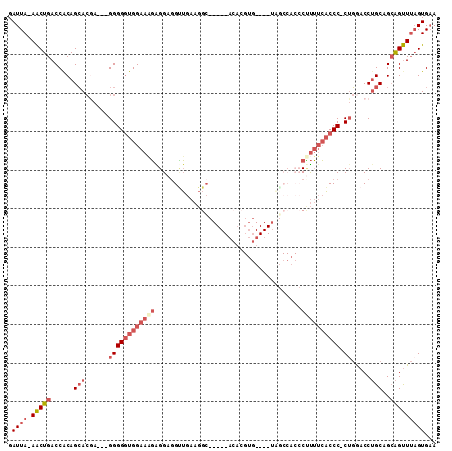
>3L_DroMel_CAF1 415256 107 - 23771897 GAUUA-AACUGACCACAGCACAA---GGGGGUGGAAUGGGGAUGUUGAAGGCCAUAUACACGUGUAUAUAGCCACCCUUUUCACCCCCUGAACCUGCAGCAGUUUAGUGAA .((((-(((((.(..(((....(---((((((((((.(((.........(((.((((((....)))))).))).))).)))))))))))....)))..))))))))))... ( -46.20) >DroGri_CAF1 73967 100 - 1 GAUUACAGCUGACCACAGGAAAAAAGGGGGCUGGCAUUGGGAGGGGGAAAGG-----ACACGCA----GUCCCACCCACAAACCCCUCUCAACCAGCUGAGGCU--GUGCA ...(((((((..((...........)).((((((..((((((((((....((-----((.....----))))..........))))))))))))))))..))))--))).. ( -39.34) >DroSec_CAF1 62175 107 - 1 GAUUA-AACUGACCACAGCACGA---GGGGGUGGAAAGAGGAGGUUGAAGGCCAUAUACACGUGUAUAUAGCCACCCUUUUCACCCACUGGACCUGCCGCAGUUUAGUGAA .((((-(((((....(((..(.(---(.((((((((((....(((....(((.((((((....)))))).)))))))))))))))).)).)..)))...)))))))))... ( -39.80) >DroSim_CAF1 64310 107 - 1 GAUUA-AACUGACCACAGCACGA---GGGGGUGGAAAGAGGAGGCUGAAGGCCAUAUACACGUGUAUAUAGCCACCCUUUUCACCCCCUGGACCUGCAGCAGUUUAGUGAA .((((-(((((.(..(((..(.(---((((((((((((.((.((((.......((((((....)))))))))).))))))))))))))).)..)))..))))))))))... ( -49.21) >DroEre_CAF1 66427 82 - 1 GAUUA-AACUGACCGCAGCACGA---GGGGGUGGAAAGAGGAGGUUGAAGGC------------------------CCUUUCACCC-CUGGACCUGCAGCAGUUUAGUGAA .((((-(((((.(.((((..(.(---((((...(((((.((..........)------------------------)))))).)))-)).)..)))).))))))))))... ( -33.50) >DroYak_CAF1 66602 81 - 1 GAUUA-AACUGACCACAGCACGA---GGGGGUGGAAAGAGGAGGUUGAAGGC------------------------CUUUUCACCC-C-GGACCUGCAGCAGUUUAGUGAA .((((-(((((.(..(((.....---((((.(....)((((((((.....))------------------------)))))).)))-)-....)))..))))))))))... ( -27.60) >consensus GAUUA_AACUGACCACAGCACGA___GGGGGUGGAAAGAGGAGGUUGAAGGC_____ACACGUG____UAGCCACCCUUUUCACCC_CUGGACCUGCAGCAGUUUAGUGAA .((((.(((((......(((......((((((((((((......................................)))))))))).)).....)))..)))))))))... (-11.66 = -13.58 + 1.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:25 2006