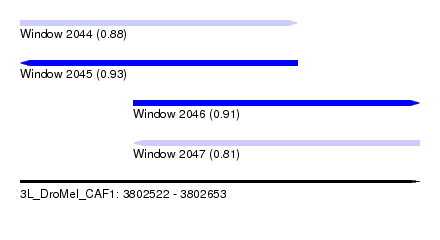
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,802,522 – 3,802,653 |
| Length | 131 |
| Max. P | 0.932307 |

| Location | 3,802,522 – 3,802,613 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -19.66 |
| Energy contribution | -21.10 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3802522 91 + 23771897 GAUGUGAACAUAAUAAUAUUAGUG--CGCUUAUUAUUACUAUUGCAACAUUUUCGGUUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUAAAGUG ..((..(...((((((((..((..--..))))))))))...)..)).((((((..(....(((((((((.....))))))))).)..)))))) ( -23.40) >DroSec_CAF1 2381 91 + 1 GAUGUGAACUUAAUAAUAUUAGUG--CGCUUAUUAUUACUAUUGCAACAUUUACGGAUCUUGCAGCCGCCGGUUGCGGUUGCAUUUUUAAGUG ..((..(...((((((((..((..--..))))))))))...)..)).((((((..((...(((((((((.....)))))))))))..)))))) ( -24.20) >DroSim_CAF1 2347 91 + 1 GAUGUGAACUUAAUAAUAUAAGUG--UGCUUAUUAUUACUAUUGCAACAUUUACGGAUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUUAAGUG .......((((((.(((.((((((--(((..............)).)))))))........((((((((.....))))))))))).)))))). ( -24.54) >DroEre_CAF1 2615 91 + 1 GAUGUGGAUUUGAAAUUAUUAGUG--CGCUUAUUAUAACGAUUGCAACAUUUUCGGUUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUUAAGUG ........................--((((((......((((((((((.....(((((.....)))))...))))))))))......)))))) ( -21.40) >DroYak_CAF1 2637 93 + 1 GAUGUGGAUUGUAAAUGAAUAGUGUGUGCUUAUUACUAUAAUUGCAACAUUUUCGGUUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUAAAGUG ........((((((....((((((.........))))))..))))))((((((..(....(((((((((.....))))))))).)..)))))) ( -23.10) >consensus GAUGUGAACUUAAUAAUAUUAGUG__CGCUUAUUAUUACUAUUGCAACAUUUUCGGUUUUUGCAGCCGCCGGUUGCGGUUGCAUUUUUAAGUG ..((..(...((((((((..(((....)))))))))))...)..)).((((((.((....(((((((((.....))))))))).)).)))))) (-19.66 = -21.10 + 1.44)


| Location | 3,802,522 – 3,802,613 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -18.12 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3802522 91 - 23771897 CACUUUAAAAUGCAACCGCAACCGGCGGCUGCAAAAACCGAAAAUGUUGCAAUAGUAAUAAUAAGCG--CACUAAUAUUAUUAUGUUCACAUC ..........((((.((((.....)))).))))..........((((.(((.((((((((.((....--...)).)))))))))))..)))). ( -18.10) >DroSec_CAF1 2381 91 - 1 CACUUAAAAAUGCAACCGCAACCGGCGGCUGCAAGAUCCGUAAAUGUUGCAAUAGUAAUAAUAAGCG--CACUAAUAUUAUUAAGUUCACAUC .((((((...((((.((((.....)))).)))).....(((...((((((....))))))....)))--...........))))))....... ( -19.50) >DroSim_CAF1 2347 91 - 1 CACUUAAAAAUGCAACCGCAACCGGCGGCUGCAAAAUCCGUAAAUGUUGCAAUAGUAAUAAUAAGCA--CACUUAUAUUAUUAAGUUCACAUC .((((((...((((.((((.....)))).))))...........((((((....))))))(((((..--..)))))....))))))....... ( -19.60) >DroEre_CAF1 2615 91 - 1 CACUUAAAAAUGCAACCGCAACCGGCGGCUGCAAAAACCGAAAAUGUUGCAAUCGUUAUAAUAAGCG--CACUAAUAAUUUCAAAUCCACAUC ..........((((.((((.....)))).))))......((((.(((((....((((......))))--...))))).))))........... ( -18.00) >DroYak_CAF1 2637 93 - 1 CACUUUAAAAUGCAACCGCAACCGGCGGCUGCAAAAACCGAAAAUGUUGCAAUUAUAGUAAUAAGCACACACUAUUCAUUUACAAUCCACAUC ..........((((.((((.....)))).))))......(((..((((((..((((....))))))).)))...)))................ ( -15.40) >consensus CACUUAAAAAUGCAACCGCAACCGGCGGCUGCAAAAACCGAAAAUGUUGCAAUAGUAAUAAUAAGCG__CACUAAUAUUAUUAAGUUCACAUC ..........((((.((((.....)))).))))...........((((((....))))))................................. (-13.28 = -13.52 + 0.24)



| Location | 3,802,559 – 3,802,653 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.87 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -13.32 |
| Energy contribution | -15.35 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3802559 94 + 23771897 UAUUGCAACAUUU------UCGGUUUUUGCAGCCGCCGG---UUGCGGUUGC-------AUUUUAAAGUGCACUACAUACGCACACACACACAUGUGCACAUACAAAUUU ...((((.(((..------........(((((((((...---..))))))))-------).......((((.........))))........)))))))........... ( -24.70) >DroSec_CAF1 2418 88 + 1 UAUUGCAACAUUU------ACGGAUCUUGCAGCCGCCGG---UUGCGGUUGC-------AUUUUUAAGUGCACUACAUA------CACACACAUGUGCACAUACAAAUUU .............------........(((((((((...---..))))))))-------).......((((((......------.........)))))).......... ( -24.26) >DroSim_CAF1 2384 88 + 1 UAUUGCAACAUUU------ACGGAUUUUGCAGCCGCCGG---UUGCGGUUGC-------AUUUUUAAGUGCACUACAUA------CACACACAUGUGCACAUACAAAUUU .............------........(((((((((...---..))))))))-------).......((((((......------.........)))))).......... ( -24.26) >DroEre_CAF1 2652 86 + 1 GAUUGCAACAUUU------UCGGUUUUUGCAGCCGCCGG---UUGCGGUUGC-------AUUUUUAAGUGCACUGCGUA------CACACACAUGU--ACAUACAUAUUU ....(((.(((((------........(((((((((...---..))))))))-------).....)))))...)))(((------((......)))--)).......... ( -24.02) >DroYak_CAF1 2676 88 + 1 AAUUGCAACAUUU------UCGGUUUUUGCAGCCGCCGG---UUGCGGUUGC-------AUUUUAAAGUGCACUACAUA------CACACACAUGUGCACAUACAUAUUU .............------........(((((((((...---..))))))))-------).......((((((......------.........)))))).......... ( -24.26) >DroAna_CAF1 2675 101 + 1 AAAGGCAGCGUUUGGGUCGCCGGUUCUUCCGGGAGUCGGAAUUGGCGGAGACACGGGAGAUUCUGGAGCGCCGGCUGUA------C-CACACAUGU--ACUUACAUAUGU ...(((.((((((((((((((((((((..(....)..)))))))))(....)......)))))..))))))..)))...------.-.(((.((((--....)))).))) ( -33.50) >consensus UAUUGCAACAUUU______UCGGUUUUUGCAGCCGCCGG___UUGCGGUUGC_______AUUUUUAAGUGCACUACAUA______CACACACAUGUGCACAUACAAAUUU ............................((((((((........))))))))...............((((((.....................)))))).......... (-13.32 = -15.35 + 2.03)



| Location | 3,802,559 – 3,802,653 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.87 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -14.03 |
| Energy contribution | -15.62 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3802559 94 - 23771897 AAAUUUGUAUGUGCACAUGUGUGUGUGUGCGUAUGUAGUGCACUUUAAAAU-------GCAACCGCAA---CCGGCGGCUGCAAAAACCGA------AAAUGUUGCAAUA ......(((((..(((((....)))))..)))))....(((.........(-------(((.((((..---...)))).))))..(((...------....))))))... ( -27.30) >DroSec_CAF1 2418 88 - 1 AAAUUUGUAUGUGCACAUGUGUGUG------UAUGUAGUGCACUUAAAAAU-------GCAACCGCAA---CCGGCGGCUGCAAGAUCCGU------AAAUGUUGCAAUA ....(((((.(((((((((((....------))))).)))))).......(-------(((.((((..---...)))).))))........------......))))).. ( -24.30) >DroSim_CAF1 2384 88 - 1 AAAUUUGUAUGUGCACAUGUGUGUG------UAUGUAGUGCACUUAAAAAU-------GCAACCGCAA---CCGGCGGCUGCAAAAUCCGU------AAAUGUUGCAAUA ....(((((.(((((((((((....------))))).)))))).......(-------(((.((((..---...)))).))))........------......))))).. ( -24.30) >DroEre_CAF1 2652 86 - 1 AAAUAUGUAUGU--ACAUGUGUGUG------UACGCAGUGCACUUAAAAAU-------GCAACCGCAA---CCGGCGGCUGCAAAAACCGA------AAAUGUUGCAAUC .....(((((((--((.(((((...------.))))))))))........(-------(((.((((..---...)))).))))........------......))))... ( -23.10) >DroYak_CAF1 2676 88 - 1 AAAUAUGUAUGUGCACAUGUGUGUG------UAUGUAGUGCACUUUAAAAU-------GCAACCGCAA---CCGGCGGCUGCAAAAACCGA------AAAUGUUGCAAUU .....((((.(((((((((((....------))))).)))))).......(-------(((.((((..---...)))).))))........------......))))... ( -23.70) >DroAna_CAF1 2675 101 - 1 ACAUAUGUAAGU--ACAUGUGUG-G------UACAGCCGGCGCUCCAGAAUCUCCCGUGUCUCCGCCAAUUCCGACUCCCGGAAGAACCGGCGACCCAAACGCUGCCUUU ((((((((....--))))))))(-(------..((((.(((((.............)))))..((((..(((((.....))))).....))))........))))))... ( -29.12) >consensus AAAUAUGUAUGUGCACAUGUGUGUG______UAUGUAGUGCACUUAAAAAU_______GCAACCGCAA___CCGGCGGCUGCAAAAACCGA______AAAUGUUGCAAUA .....((((.((((((.((((((........))))))))))))...............(((.((((........)))).))).....................))))... (-14.03 = -15.62 + 1.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:28 2006