
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,780,384 – 3,780,509 |
| Length | 125 |
| Max. P | 0.619099 |

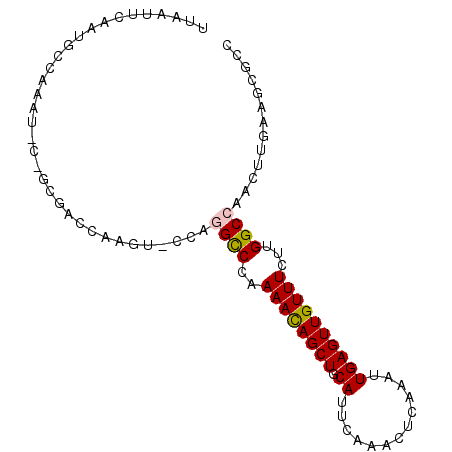
| Location | 3,780,384 – 3,780,478 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 83.48 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -11.94 |
| Energy contribution | -12.02 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
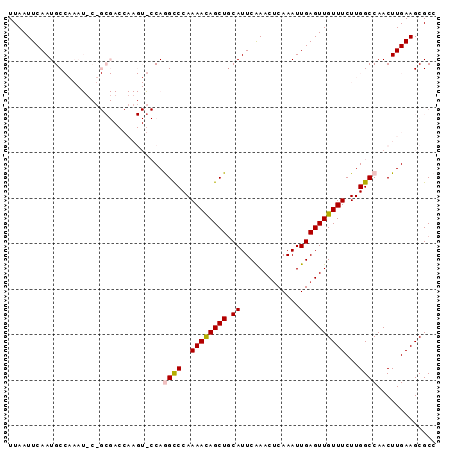
>3L_DroMel_CAF1 3780384 94 + 23771897 UUAAUUCAAUGCCAAAUUCGGCGACCAAGUACCAGGCCCAAAAUAGCUGCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCC ...................((((..(((((....((((..((((((((.((.............))))))))))...)))).)))))...)))) ( -22.92) >DroSec_CAF1 177983 74 + 1 UUAAUUCAAUGC--------------------CAGGCCCAAAACAGCUUCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCC ....(((((.((--------------------((((....(((((((((.(((........))).)))))))))))))))....)))))..... ( -17.80) >DroSim_CAF1 172343 74 + 1 UUAAUUCAAUGC--------------------CAGGCCCAAAACAGCUGCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCC ....(((((.((--------------------((((....((((((((.((.............))))))))))))))))....)))))..... ( -16.62) >DroEre_CAF1 170821 94 + 1 UUAAUUCAAUGCCAAAUUCUGCGACCAAGUGCCAAGCCCAAAACAGCUGCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCC ....................(((..(((((((((((....((((((((.((.............))))))))))))))))..)))))...))). ( -23.42) >DroYak_CAF1 162915 94 + 1 UUAAUUCAAUGCCAAAUGCUGCGACCAAGUGCCAAGUCCAAAACAGCUGCAUUCGAACGCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCC ....................(((..(((((((((((....((((((((.((.............))))))))))))))))..)))))...))). ( -23.42) >consensus UUAAUUCAAUGCCAAAU_C_GCGACCAAGU_CCAGGCCCAAAACAGCUGCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCC ..................................((((..((((((((.((.............))))))))))...))))............. (-11.94 = -12.02 + 0.08)

| Location | 3,780,415 – 3,780,509 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 91.89 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -17.34 |
| Energy contribution | -17.42 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
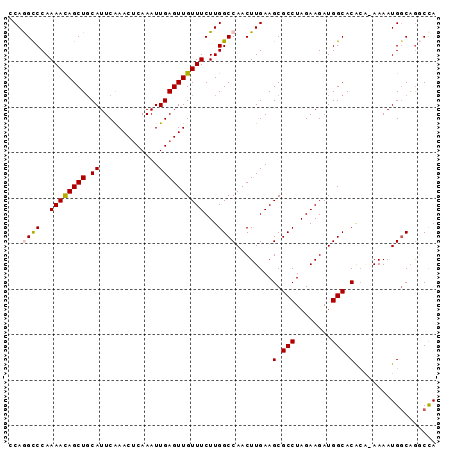
>3L_DroMel_CAF1 3780415 94 + 23771897 CCAGGCCCAAAAUAGCUGCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCCUAGAAGAUGGCACACAGAAAAUGUCAGGCCA ...((((........(((......(((((.....)))))(((((..(.....)..)))))(((........)))...))).........)))). ( -24.23) >DroSec_CAF1 177995 93 + 1 -CAGGCCCAAAACAGCUUCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCCUAGAAGAUGGCACACAAAAAAUGGCAGGCUA -..((((..(((((((((.(((........))).)))))))))....((((..(((..(.(((........))).).)))....)))).)))). ( -23.40) >DroSim_CAF1 172355 93 + 1 -CAGGCCCAAAACAGCUGCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCCUAGAAGAUGGCACACAAAAAAUGGCAGGCCA -..((((..((((((((.((.............))))))))))....((((..(((..(.(((........))).).)))....)))).)))). ( -24.82) >DroEre_CAF1 170852 92 + 1 CCAAGCCCAAAACAGCUGCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCCUAGAAGAUGGCACAC--AAAAUGGCCACCCA .........((((((((.((.............))))))))))..((((((..(((..(.(((........))).).)--))..)))))).... ( -22.22) >DroYak_CAF1 162946 92 + 1 CCAAGUCCAAAACAGCUGCAUUCGAACGCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCCUAGAAGAUGGCACGA--AAAAUGGCAGCCCA ..............(((((.((((...((...(..(((((.........)))))..).))(((........))).)))--).....)))))... ( -23.00) >consensus CCAGGCCCAAAACAGCUGCAUUCAAACUCAAAUUGAGUUGUUUCUUGGCCAACUUGAAGCGCCUAGAAGAUGGCACACA_AAAAUGGCAGGCCA ...((((..((((((((.((.............))))))))))...))))........(.(((........))).).................. (-17.34 = -17.42 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:17 2006