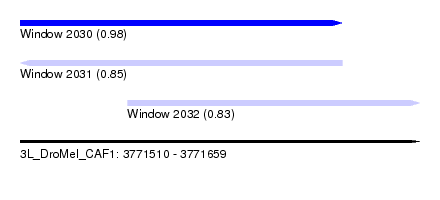
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,771,510 – 3,771,659 |
| Length | 149 |
| Max. P | 0.975257 |

| Location | 3,771,510 – 3,771,630 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -43.58 |
| Consensus MFE | -36.20 |
| Energy contribution | -36.88 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
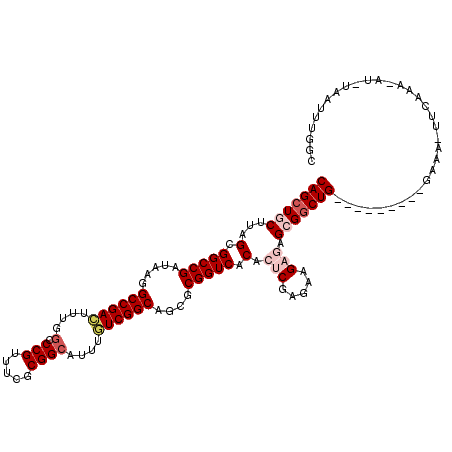
>3L_DroMel_CAF1 3771510 120 + 23771897 AAAAUUGUGACAAAAAAUUGUUUAUAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGAAUUUGCACCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCGAGAAGAGA .....((((((((....))).)))))...((.(((((((((.(((((....(((((.....)))))................((((....)))))))))..)))))))))))........ ( -42.90) >DroSec_CAF1 169017 120 + 1 AAAUUUGUGACAAAAAAUUGUUUAUAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGACAUUGGCCCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCGAGAAGAGA .....((((((((....))).)))))...((.(((((((((.((((((....((((.....))))((((...(((((...)).)))...))))))))))..)))))))))))........ ( -48.60) >DroSim_CAF1 163401 120 + 1 AAAUUUGUGACAAAAAAUUGUUUAUAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGACUUUGGCCCGUUUCGCGGCACUUGUCGGCAGCGCGGUCACACUCGAGAAGAGA .....((((((((....))).)))))...((.(((((((((.((((((....((((.....))))(((....(((((...)).)))....)))))))))..)))))))))))........ ( -46.90) >DroEre_CAF1 161718 109 + 1 -AAAUUGUGACAAAAAAUUGUUUAUAUGGCGCAGUGUGGCCAGAUACUUGGCGGCCGAUAAGGCCGAGUUGGGCCCGUUUGGCGGCAUUUUUCGGCAGCGCGGUCACACC---------- -....((((((........(((.....)))...((((.(((.((......((.(((((...((((......))))...))))).)).....))))).)))).))))))..---------- ( -39.50) >DroYak_CAF1 153126 110 + 1 AAAAUUGUGACAAAAAAUUGUUUAUAUGGCGCAGUGUGGCCAGCUACUAGGCGGCCGAUGAGGCCGAGUUUCGCCCGUUUGGCGGCAUUCUUCGGCAGCGCGGUCACACC---------- .....((((((........(((.....)))...((((.(((.(((....)))((((.....))))((((.(((((.....))))).))))...))).)))).))))))..---------- ( -40.00) >consensus AAAAUUGUGACAAAAAAUUGUUUAUAUGCCGCAGUGUGGCCAGCUGCUUAGCGGCCGAUAAGGCCGACUUUGGCCCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCGAGAAGAGA .....((((.((..............)).))))((((((((.((((((....((((.....))))(((....(.(((.....))))....)))))))))..))))))))........... (-36.20 = -36.88 + 0.68)



| Location | 3,771,510 – 3,771,630 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -31.38 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3771510 120 - 23771897 UCUCUUCUCGAGUGUGACCGCGCUGCCGACAAAUGCCGCGAAACGGUGCAAAUUCGGCCUUAUCGGCCGCUAAGCAGCUGGCCACACUGCGGCAUAUAAACAAUUUUUUGUCACAAUUUU .......(((((((((.(((.(((((........((((.....)))).......(((((.....)))))....)))))))).)))))).))).......((((....))))......... ( -36.70) >DroSec_CAF1 169017 120 - 1 UCUCUUCUCGAGUGUGACCGCGCUGCCGACAAAUGCCGCGAAACGGGCCAAUGUCGGCCUUAUCGGCCGCUAAGCAGCUGGCCACACUGCGGCAUAUAAACAAUUUUUUGUCACAAAUUU .(((.....)))((((((((......))....((((((((.....(((((.((((((((.....)))))....)))..)))))....))))))))..............))))))..... ( -41.60) >DroSim_CAF1 163401 120 - 1 UCUCUUCUCGAGUGUGACCGCGCUGCCGACAAGUGCCGCGAAACGGGCCAAAGUCGGCCUUAUCGGCCGCUAAGCAGCUGGCCACACUGCGGCAUAUAAACAAUUUUUUGUCACAAAUUU .(((.....)))((((((((......))....(((((((((...(((((......)))))..))(((((((....))).)))).....)))))))..............))))))..... ( -41.10) >DroEre_CAF1 161718 109 - 1 ----------GGUGUGACCGCGCUGCCGAAAAAUGCCGCCAAACGGGCCCAACUCGGCCUUAUCGGCCGCCAAGUAUCUGGCCACACUGCGCCAUAUAAACAAUUUUUUGUCACAAUUU- ----------..((((((.(.((.(((((.......((.....))((((......))))...))))).)))...(((.(((((.....).)))).)))...........))))))....- ( -31.60) >DroYak_CAF1 153126 110 - 1 ----------GGUGUGACCGCGCUGCCGAAGAAUGCCGCCAAACGGGCGAAACUCGGCCUCAUCGGCCGCCUAGUAGCUGGCCACACUGCGCCAUAUAAACAAUUUUUUGUCACAAUUUU ----------((((((.(((.(((((..........((((.....)))).....(((((.....)))))....)))))))).))))))...........((((....))))......... ( -34.50) >consensus UCUCUUCUCGAGUGUGACCGCGCUGCCGACAAAUGCCGCGAAACGGGCCAAACUCGGCCUUAUCGGCCGCUAAGCAGCUGGCCACACUGCGGCAUAUAAACAAUUUUUUGUCACAAUUUU ..........((((((.(((.(((((.........(((.....)))........(((((.....)))))....)))))))).))))))...........((((....))))......... (-31.38 = -31.10 + -0.28)


| Location | 3,771,550 – 3,771,659 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -41.04 |
| Consensus MFE | -29.24 |
| Energy contribution | -30.82 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3771550 109 + 23771897 CAGCUGCUUAGCGGCCGAUAAGGCCGAAUUUGCACCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCGAGAAGAGAGCGGCUG---------GAAA-UUCAAA-AUUUAAUUUGGC ((((((((..(.(((((.....(((((...(((..((...))..)))....)))))....))))).).(((.....)))))))))))---------....-.(((((-......))))). ( -39.80) >DroSec_CAF1 169057 110 + 1 CAGCUGCUUAGCGGCCGAUAAGGCCGACAUUGGCCCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCGAGAAGAGAGCGGCUG---------GAAA-UUCAAAAAUGUAAUUUGGC ((((((((..(.(((((.....(((((((...(((((...)).)))...)))))))....))))).).(((.....)))))))))))---------....-................... ( -45.10) >DroSim_CAF1 163441 109 + 1 CAGCUGCUUAGCGGCCGAUAAGGCCGACUUUGGCCCGUUUCGCGGCACUUGUCGGCAGCGCGGUCACACUCGAGAAGAGAGCGGCUG---------GAAA-UUCAAA-AUUUAAUUUGGC ((((((((..(.(((((.....((((((....(((((...)).)))....))))))....))))).).(((.....)))))))))))---------....-.(((((-......))))). ( -43.90) >DroEre_CAF1 161757 104 + 1 CAGAUACUUGGCGGCCGAUAAGGCCGAGUUGGGCCCGUUUGGCGGCAUUUUUCGGCAGCGCGGUCACACC----------GCGGCUGUAGUGAGCGGAAU-UUCAAA-AU----UUUGGC ....((((..((.(((((...((((......))))...))))).))........(((((((((.....))----------)).)))))))))..((((((-(....)-))----)))).. ( -38.20) >DroYak_CAF1 153166 104 + 1 CAGCUACUAGGCGGCCGAUGAGGCCGAGUUUCGCCCGUUUGGCGGCAUUCUUCGGCAGCGCGGUCACACC----------GCGGCUGUA-UGAAUGAAAUUUUCAAA-AU----UUUGGC ..((((.....(((((.....)))))....(((((.....)))))(((((....(((((((((.....))----------)).))))).-.)))))...........-..----..)))) ( -38.20) >consensus CAGCUGCUUAGCGGCCGAUAAGGCCGACUUUGGCCCGUUUCGCGGCAUUUGUCGGCAGCGCGGUCACACUCGAGAAGAGAGCGGCUG_________GAAA_UUCAAA_AU_UAAUUUGGC (((((((...(.(((((.....((((((....(.(((.....))))....))))))....))))).).(((.....))).)))))))................................. (-29.24 = -30.82 + 1.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:14 2006