| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 413,446 – 413,549 |
| Length | 103 |
| Max. P | 0.841270 |

| Location | 413,446 – 413,549 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.25 |
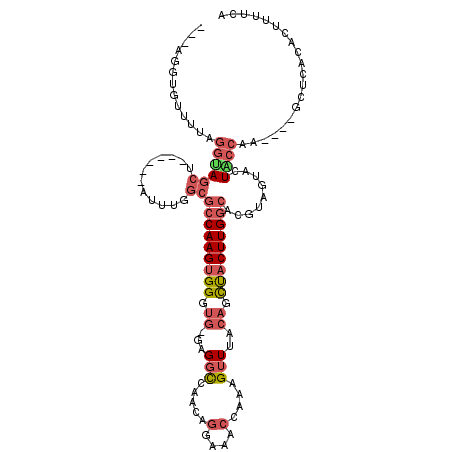
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -15.79 |
| Energy contribution | -17.10 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
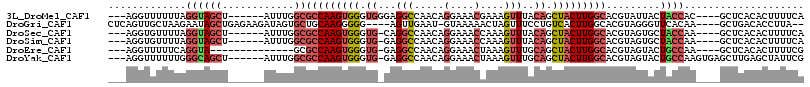
>3L_DroMel_CAF1 413446 103 + 23771897 ---AGGUUUUUUAGGUAGCU------AUUUGGCGCCAAGUGGGUGGGAGGCCAACAGGAAACGAAAGUUUACAGCUACUUGGCACGUAUUACUACCAC----GCUCACACUUUUCA ---..........(((((..------((.((..(((((((((.((.....((....))((((....)))).)).))))))))).)).))..)))))..----.............. ( -32.10) >DroGri_CAF1 71914 105 + 1 CUCAGUUGCUAAGAAUAGCUGAGAAGAUAGUGCUGCAAGGGGG----AGUUGAAU-GUAAAAACUAGUUUCCUGUCACUUGGCACGUAGGGUUCACAA----GCUGACACCUUA-- ((((((((.......))))))))......((((((((.((..(----((((....-.....))))..)..))))).....))))).(((((((((...----..))).))))))-- ( -26.30) >DroSec_CAF1 60329 102 + 1 ---AGGUGUUUUAGGUAGCU------AUUUGGCGCCAAGUGGGUG-CAGGCCAACAGGAAACCAAAGUUUACAGCUACUUGGCACGUAGUGCUACCAA----GCUCACACUUUUCA ---..(((..((.((((((.------...((..(((((((((.((-.((((.....(....)....)))).)).))))))))).))....)))))).)----)..)))........ ( -35.80) >DroSim_CAF1 62452 102 + 1 ---AGGUGUUUUAGGUAGCU------AUUUGGCGCCAAGUGGGUG-GAGGCCAACAGGAAACCAAAGUUUACAGCUACUUGGCACGUAGUGCUACCAA----GCUCACACUUUUCA ---..(((..((.((((((.------...((..(((((((((.((-.((((.....(....)....)))).)).))))))))).))....)))))).)----)..)))........ ( -35.80) >DroEre_CAF1 64618 94 + 1 ---AGGUUUUUCAGGUA--------------GCGCCAAGUGGGUG-GAGGCCAACAGGAAACUAAAGUUUGCAGCUACUUGGCACGUAGUACUGCCAA----GCUCACACUUUUCG ---.(((((..(((...--------------(.(((((((((.((-.((((....((....))...)))).)).))))))))).)......)))..))----)))........... ( -30.30) >DroYak_CAF1 64693 106 + 1 ---AGGUUUUUUGGGCAGCU------AUUUGGCGCCAAGUGGGUG-GAGGCCAACAGGAAACUAAAGUUUGCAGCUACUUGGCACGUAGUACUGCCAAGUGAGCUUGAGCUAUUCG ---((((((....(((((.(------(((..(.(((((((((.((-.((((....((....))...)))).)).))))))))).)..)))))))))....)))))).......... ( -40.50) >consensus ___AGGUGUUUUAGGUAGCU______AUUUGGCGCCAAGUGGGUG_GAGGCCAACAGGAAACCAAAGUUUACAGCUACUUGGCACGUAGUACUACCAA____GCUCACACUUUUCA .............((((((............))(((((((((.((...(((.....(....)....)))..)).))))))))).........)))).................... (-15.79 = -17.10 + 1.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:20 2006