
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,737,826 – 3,737,968 |
| Length | 142 |
| Max. P | 0.633223 |

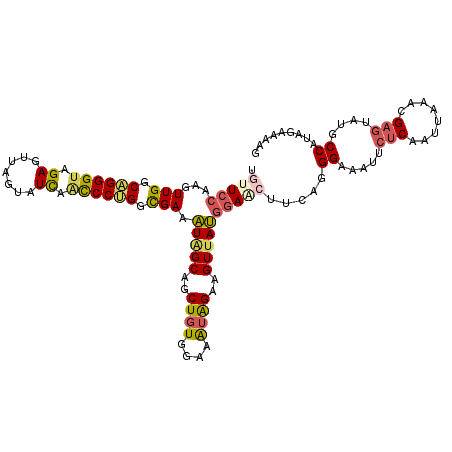
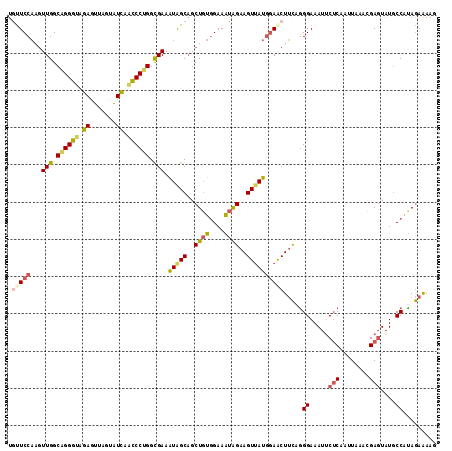
| Location | 3,737,826 – 3,737,941 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.97 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -24.29 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3737826 115 - 23771897 UGUUCCAAGUUGGCAGGGUAGAGUUAGUAUCAACCCUGGCGAAAUGGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGGGAAAUUCUCAAUUAAACGAGUAUGCCGUAGAAAAG .(((((...(((.((((((.((.......)).)))))).))).(((((..((((....))))..))))))))))((((.((.....(((........)))....)).).)))... ( -34.50) >DroSec_CAF1 132630 115 - 1 UGUUCCAAGUUGGCAGGGUAGAGUUAGUAUCAACCCUGGCGAAAUAGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGGGAAAUUCUCAAUUAAACGAGUAUGCCAUAGAAAAG .(((((...(((.((((((.((.......)).)))))).))).(((((..((((....))))..))))))))))(((..((.....(((........)))....))...)))... ( -34.40) >DroSim_CAF1 128842 115 - 1 UGCUCCAAGUUGGCAGGGUAGAGUUAGUAUCAACCCUGGCGAAAUAGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGGGAAAUUCUCAAUUAAACGAGUAUGCCAUAGAAAAG ((((.....(((.((((((.((.......)).)))))).)))...)))).((((((.....((((((....)))))).(((.....)))...............))))))..... ( -33.90) >DroEre_CAF1 127870 115 - 1 UACUCCAAGUUGGCUGGGUAGAAUUAGUAUCAGCCCUGGCGAAAUAGCAGCUGUGGAAGUAGAAGUUAUGGAACUUCAGGGAAAUUCUCAAUUAAACGAGUAUGCCACAGAAAAG .........((.(((((((.((.......)).)))).))).)).......((((((.....((((((....)))))).(((.....)))...............))))))..... ( -27.50) >DroYak_CAF1 116613 115 - 1 UGUUCCAAGUUGGCAGGGUAGAAUUAGUAUUAACCCUGGCGAAAUAGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGGGAAAUGCUCAACUAAACGAGUUUGCCGUAGAGAAG .(((((...(((.((((((..(((....))).)))))).))).(((((..((((....))))..))))))))))((((.((....((((........))))...)).).)))... ( -34.20) >DroAna_CAF1 130917 115 - 1 UAUUCCUAAUUGGCUGGGCAGAGUUAGUGUUAAUCCAGGUGAAGUGGCCGCUCUAGAAGUGGAAGUAACUCAGCUAUUAGGAAAUUCACAACUUAAGGAUUAUUCCAUAAGAAAA ..((((((((.(((((((................(((.......)))(((((.....)))))......)))))))))))))))........((((.(((....))).)))).... ( -30.80) >consensus UGUUCCAAGUUGGCAGGGUAGAGUUAGUAUCAACCCUGGCGAAAUAGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGGGAAAUUCUCAAUUAAACGAGUAUGCCAUAGAAAAG .(((((...(((.((((((.((.......)).)))))).))).(((((..((((....))))..)))))))))).....((.....(((........)))....))......... (-24.29 = -24.63 + 0.34)

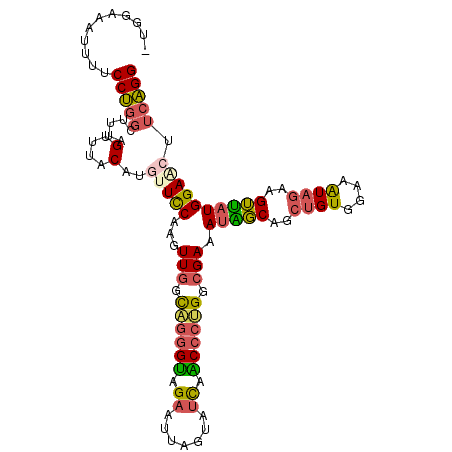
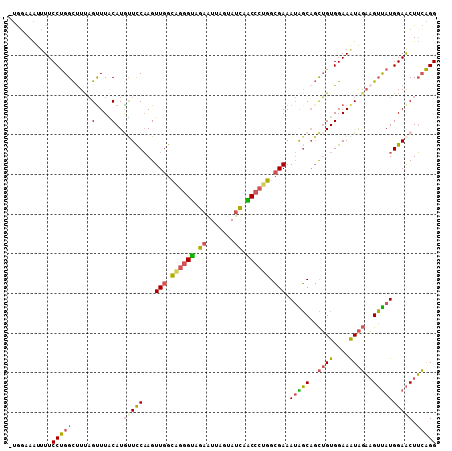
| Location | 3,737,861 – 3,737,968 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -21.97 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3737861 107 - 23771897 -UGGAAAUCUUCCUGGCUUUAGUUUACAUGUUCCAAGUUGGCAGGGUAGAGUUAGUAUCAACCCUGGCGAAAUGGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGG -..(((...(((((((((((..((((((.((((((..(((.((((((.((.......)).)))))).)))..))).)))))))))....))))))).)))).)))... ( -36.10) >DroPse_CAF1 176291 108 - 1 AGAGCAGUUUUCCCGGUUUCCGUUUGCAUAUUUCCAGUUGGUUGGGCCGCAUAACCAUCGGCGGGGGGGAAAAGUCCCUGGUGGAAGUGCUCGGCAUGGAGCUGGGGG ..........((((((((.(((((((..(((((((.(.((((((.......)))))).).((.(((((......))))).)))))))))..))).)))))))))))). ( -38.90) >DroSec_CAF1 132665 107 - 1 -UGGAAAUUUUCCUGGCUUUAGUUUACAUGUUCCAAGUUGGCAGGGUAGAGUUAGUAUCAACCCUGGCGAAAUAGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGG -..........((((((....))......(((((...(((.((((((.((.......)).)))))).))).(((((..((((....))))..))))))))))..)))) ( -32.30) >DroSim_CAF1 128877 107 - 1 -UGGAAAUUUUCCUGGCUUUAGUUUGCAUGCUCCAAGUUGGCAGGGUAGAGUUAGUAUCAACCCUGGCGAAAUAGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGG -..(((...(((((((((((.((((.(((((((....(((.((((((.((.......)).)))))).)))....).))).))).)))).))))))).)))).)))... ( -34.20) >DroEre_CAF1 127905 107 - 1 -UGACAAUUUUCCUGGCUUUAGUUUGCAUACUCCAAGUUGGCUGGGUAGAAUUAGUAUCAGCCCUGGCGAAAUAGCAGCUGUGGAAGUAGAAGUUAUGGAACUUCAGG -........(((((((((((..(((.((((((.(...((.(((((((.((.......)).)))).))).))...).)).)))).)))..))))))).))))....... ( -26.70) >DroYak_CAF1 116648 107 - 1 -UGGUAAUUUUCCUGGCCUCAGCUUACAUGUUCCAAGUUGGCAGGGUAGAAUUAGUAUUAACCCUGGCGAAAUAGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGG -..........((((((....))......(((((...(((.((((((..(((....))).)))))).))).(((((..((((....))))..))))))))))..)))) ( -31.30) >consensus _UGGAAAUUUUCCUGGCUUUAGUUUACAUGUUCCAAGUUGGCAGGGUAGAAUUAGUAUCAACCCUGGCGAAAUAGCAGCUGUGGAAAUAGAAGUUAUGGAACUUCAGG ...........(((((.....(....)..(((((...(((.((((((.((.......)).)))))).))).(((((..((((....))))..)))))))))).))))) (-21.97 = -22.08 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:00 2006