| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
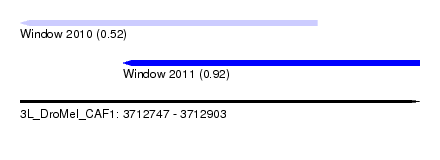
| Location | 3,712,747 – 3,712,903 |
| Length | 156 |
| Max. P | 0.922260 |

| Location | 3,712,747 – 3,712,863 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -24.90 |
| Energy contribution | -26.74 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3712747 116 - 23771897 CCAAGUUCGAAACUGCGAAUGAGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACU----ACAACAAGUGCGGGGAAAAACCGGAAACUUUACGAGCCGCA ((..(((((......)))))..))...((((.(((((((.....)))))))((....))............))))----........(((((((.....))(....)........))))) ( -32.00) >DroSec_CAF1 107810 116 - 1 CCAAGUUCGAAACUGCAAAUGGGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACU----ACAACAAGUGCGGGGAAAAACCGGAAACUUUACGAGCCGCA ....(((((...(((((..((..(...((((.(((((((.....)))))))((....))............))))----.)..))..)))))((.....))(....)....))))).... ( -32.40) >DroSim_CAF1 103534 116 - 1 CCAAGUUCGAAACUGCAAAUGGGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACU----ACAACAAGUGCGGGGAAAAAGCGGAAACUUUACGAGCCGCA ....(((((...(((((..((..(...((((.(((((((.....)))))))((....))............))))----.)..))..))))).........(....)....))))).... ( -32.30) >DroEre_CAF1 102907 120 - 1 CCAAGUUCGAAACUGCAAAUGGGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACUGGGUACAACAAGUGCGGGGAAAAAUCGGAAACUUUACGAGCCGCA ....(((((...(((((..((..((..((((.(((((((.....)))))))((....))............))))...).)..))..))))).........(....)....))))).... ( -32.80) >DroYak_CAF1 91035 100 - 1 CCAAGUUCGAAACUGCAAAUGGGG---------CU-------UUCCAAUGCGGUUUUCCAACAUAAUCAAAGACU----ACAACAAGUGCGGGGAAAAAUCGGAAACUUUACGAGCCGCA ....(((.(((((((((..((((.---------..-------.)))).))))).)))).))).............----........(((((.((....))(....)........))))) ( -20.80) >consensus CCAAGUUCGAAACUGCAAAUGGGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACU____ACAACAAGUGCGGGGAAAAACCGGAAACUUUACGAGCCGCA ....(((((...(((((..((..(...((((.(((((((.....)))))))((....))............)))).....)..))..))))).........(....)....))))).... (-24.90 = -26.74 + 1.84)



| Location | 3,712,787 – 3,712,903 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -22.80 |
| Energy contribution | -24.20 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3712787 116 - 23771897 AGAAUCGAGUCGGAAAAAAAGUUAUGCCAAAGUUGGUGAACCAAGUUCGAAACUGCGAAUGAGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACU----A .......((((((.............))...(((((.(((((..(((((......)))))............(((((((.....)))))))))))).))))).........))))----. ( -32.62) >DroSec_CAF1 107850 108 - 1 -------AGUCGGAAA-AAAGUUAUGCCAAAGUUGGUGAACCAAGUUCGAAACUGCAAAUGGGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACU----A -------((((((...-.........))...(((((.(((((.....(((..((.(....).)).....)))(((((((.....)))))))))))).))))).........))))----. ( -31.00) >DroSim_CAF1 103574 115 - 1 AGAAUCGAGUCGGAAA-AAAGUUAUGCCAAAGUUGGUGAACCAAGUUCGAAACUGCAAAUGGGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACU----A .......((((((...-.........))...(((((.(((((.....(((..((.(....).)).....)))(((((((.....)))))))))))).))))).........))))----. ( -31.40) >DroEre_CAF1 102947 120 - 1 AGAAUCGAGUCGGACAAAAAGUUAUGCCAAAGUUGGUGAACCAAGUUCGAAACUGCAAAUGGGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACUGGGUA ...(((.((((((.((........))))...(((((.(((((.....(((..((.(....).)).....)))(((((((.....)))))))))))).))))).........)))).))). ( -37.00) >DroYak_CAF1 91075 100 - 1 AGAAGCGAGUCCGCAAAAAAGUUAUGCCAAAGUUGGUGAACCAAGUUCGAAACUGCAAAUGGGG---------CU-------UUCCAAUGCGGUUUUCCAACAUAAUCAAAGACU----A .......((((.(((.........)))....(((((.((((...))))(((((((((..((((.---------..-------.)))).)))))))))))))).........))))----. ( -23.80) >consensus AGAAUCGAGUCGGAAAAAAAGUUAUGCCAAAGUUGGUGAACCAAGUUCGAAACUGCAAAUGGGGAAAGGUCGGCGGUUCAACUUGAACUGCGGUUUUCCAACAUAAUCAAAGACU____A .......((((((.............))...(((((.(((((..........((.(....).))........(((((((.....)))))))))))).))))).........))))..... (-22.80 = -24.20 + 1.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:54 2006