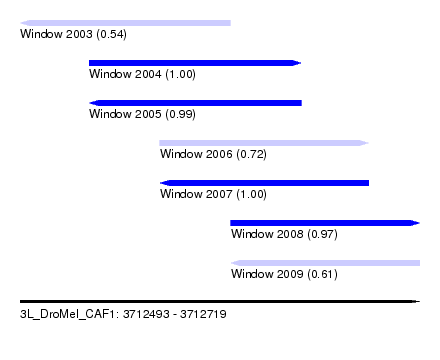
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,712,493 – 3,712,719 |
| Length | 226 |
| Max. P | 0.999645 |

| Location | 3,712,493 – 3,712,612 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.49 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3712493 119 - 23771897 AGCGACGGAGAAAGCAGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUAGGCGCAAAAGUCCUCAUCGAGAAAUGGGUAUUC-CAUUUAAUUGGAAUAUACCAC ..(((..(((..(((.......(((((..(....)..)))))....((((((...))))))))).(((......)))))).))).....(((((((((-((......)))))))).))). ( -29.90) >DroSec_CAF1 107558 120 - 1 AGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGACUGGGCACAAAAGUCCUCAUCGAGAAACGGGUAUUCCCGUUUAAUUGGAAUAUACCAC ..(.((.......)).).......(((((((......)))...(((((((((...(((.(((((.........)))))..)))...(((((((....))))))))))))))))...)))) ( -35.30) >DroSim_CAF1 103282 120 - 1 AGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUGGGCACAAAAGUCCUCAUCGAGAAACGGGUAUUCCCGUUUAAUUGGAAUAUACCAC ..(.((.......)).).......(((((((......)))...(((((((((...(((.(((((.........)))))..)))...(((((((....))))))))))))))))...)))) ( -34.70) >DroEre_CAF1 102656 119 - 1 AGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUGGGCGCAAAAGUGCUCAUCGAGAAACGGGUAUUC-CAUUUAAUUGGAAUAUACCAC ..(.((.......)).).......(((((((......)))...((((((((((((...((((.(((((((....)))))))(((.....)))....))-))))))))))))))...)))) ( -32.70) >DroYak_CAF1 90756 119 - 1 AGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUGGGCGCAAAAGUCCUCAUCGAGAAACGGGUAUUC-CAUUUAAUUGGAAUAUACCAC ..(.((.......)).).....((.(.((((((......(((...(((((((...))))))))))(((......))))))))).).))..((((((((-((......)))))))).)).. ( -30.70) >consensus AGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUGGGCGCAAAAGUCCUCAUCGAGAAACGGGUAUUC_CAUUUAAUUGGAAUAUACCAC ..(.((.......)).).......(((((((......)))...(((((((((((((((.(((((.........)))))..)))(.(((.......))).).))))))))))))...)))) (-25.04 = -25.28 + 0.24)


| Location | 3,712,532 – 3,712,652 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -28.44 |
| Energy contribution | -28.24 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3712532 120 + 23771897 GAGGACUUUUGCGCCUAGCCCAAUCUCUAAUUGGAAUUCUGUGUUUUCUCACCCACAAUUUCUCUGCUUUCUCCGUCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUA ..(((....(((((((((.(((((.....))))).....((((..........))))....................((...(((((....))))).))........)))))))))))). ( -28.60) >DroSec_CAF1 107598 120 + 1 GAGGACUUUUGUGCCCAGUCCAAUCUCUAAUUGGAAUUCUGUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUA ..(((....(((((((((((((((.....))))))....((((..........))))....................((...(((((....))))).))........)))))))))))). ( -30.70) >DroSim_CAF1 103322 120 + 1 GAGGACUUUUGUGCCCAGCCCAAUCUCUAAUUGGAAUUCUGUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGCCGUUGCCUACUUUUCUGGGCGCAUCUA (((((...(((((......(((((.....)))))......(((......))).))))).))))).((..........((.....))...........(((((......)))))))..... ( -22.20) >DroEre_CAF1 102695 120 + 1 GAGCACUUUUGCGCCCAGCCCAAUCUCUAAUUGGAAUUCUGUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUA .........(((((((((.(((((.....))))).....((((..........))))....................((...(((((....))))).))........))))))))).... ( -30.80) >DroYak_CAF1 90795 120 + 1 GAGGACUUUUGCGCCCAGCCCAAUCUCUAAUUGGAAUUCUGUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUA ..(((....(((((((((.(((((.....))))).....((((..........))))....................((...(((((....))))).))........)))))))))))). ( -30.90) >consensus GAGGACUUUUGCGCCCAGCCCAAUCUCUAAUUGGAAUUCUGUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUA .........(((((((((.(((((.....))))).....((((..........))))....................((...(((((....))))).))........))))))))).... (-28.44 = -28.24 + -0.20)

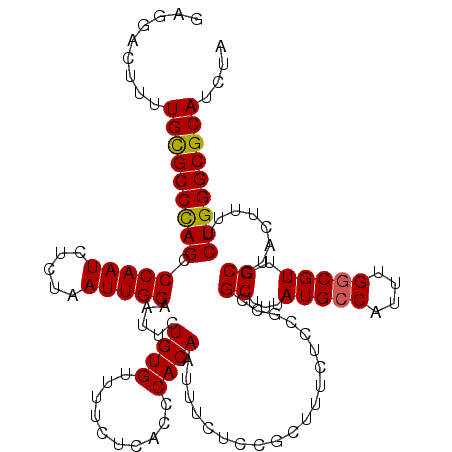
| Location | 3,712,532 – 3,712,652 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.64 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3712532 120 - 23771897 UAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGACGGAGAAAGCAGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUAGGCGCAAAAGUCCUC ....((((((.......((.(....)(((....))).....)).........(((.......(((((..(....)..)))))....((((((...))))))))).))))))......... ( -29.50) >DroSec_CAF1 107598 120 - 1 UAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGACUGGGCACAAAAGUCCUC ....((.((((((.......(....)(((....)))......(.((.......)).).....(((((..(....)..)))))...(((((((...))))))))))))).))......... ( -31.20) >DroSim_CAF1 103322 120 - 1 UAGAUGCGCCCAGAAAAGUAGGCAACGGCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUGGGCACAAAAGUCCUC .......((((((....((.(....).)).....((.....)).((.......))((((...(((((..(....)..)))))..)))).........)))))).((((......)))).. ( -27.50) >DroEre_CAF1 102695 120 - 1 UAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUGGGCGCAAAAGUGCUC ....(((((((((...(((...(..((((.....((.....))))))........((((...(((((..(....)..)))))..)))).....)..)))...)))))))))......... ( -35.40) >DroYak_CAF1 90795 120 - 1 UAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUGGGCGCAAAAGUCCUC ....(((((((((...(((...(..((((.....((.....))))))........((((...(((((..(....)..)))))..)))).....)..)))...)))))))))......... ( -35.40) >consensus UAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACACAGAAUUCCAAUUAGAGAUUGGGCUGGGCGCAAAAGUCCUC ....(((((((((....((.(....)(((....)))........))................(((((..(....)..)))))...(((((((...))))))))))))))))......... (-29.16 = -29.64 + 0.48)

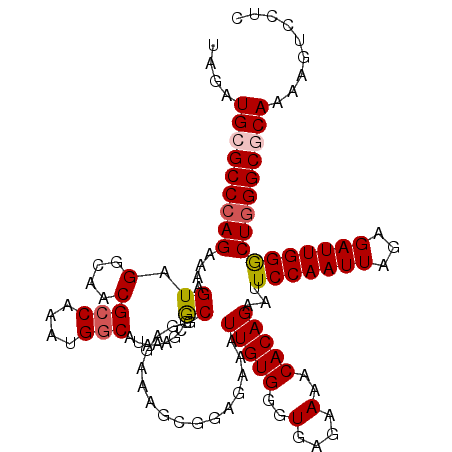
| Location | 3,712,572 – 3,712,690 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -30.78 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3712572 118 + 23771897 GUGUUUUCUCACCCACAAUUUCUCUGCUUUCUCCGUCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGGCGCGCGUG--UGUGUGGGCU ...........((((((.......(((...............(((((....))))).(((((......))))))))...........((((((((......))).)))--)))))))).. ( -33.40) >DroSec_CAF1 107638 118 + 1 GUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGGCGCGCGUG--UGUGUGGGCU ...........((((((....(..(((......(((((((..(((((....))))).(((((......)))))......................))))))).))).)--..)))))).. ( -33.60) >DroSim_CAF1 103362 118 + 1 GUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGCCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGGCGCGCGUG--UGUGUGGGCU ...........((((((........((..........((.....))...........(((((......)))))))............((((((((......))).)))--)))))))).. ( -28.50) >DroEre_CAF1 102735 118 + 1 GUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGCCGCGCGUG--UGUGUGGGCU ...........((((((........((......((((((.....)).....))))..(((((......)))))))............((((((((......))).)))--)))))))).. ( -33.40) >DroYak_CAF1 90835 120 + 1 GUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGGCGUGCGUGUGUGUGUGGGCU ...........((((((....(.((((......(((((((..(((((....))))).(((((......)))))......................))))))).))).).)..)))))).. ( -35.80) >consensus GUGUUUUCUCACCCACAAUUUCUCCGCUUUCUCCGCCGCUUUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGGCGCGCGUG__UGUGUGGGCU ...........((((((.......(((......(((((((..(((((....))))).(((((......)))))......................))))))).)))......)))))).. (-30.78 = -30.90 + 0.12)



| Location | 3,712,572 – 3,712,690 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -32.76 |
| Energy contribution | -33.56 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3712572 118 - 23771897 AGCCCACACA--CACGCGCGCCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGACGGAGAAAGCAGAGAAAUUGUGGGUGAGAAAACAC .((((((...--..(((((((((((..((((((...........))))))......))).)))...(((....))).....))).)).........((....)))))))).......... ( -33.90) >DroSec_CAF1 107638 118 - 1 AGCCCACACA--CACGCGCGCCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACAC .(((((((..--..(((.((((.(((.((((((...........)))))))))....((.(....)(((....))).....))))))......))).......))))))).......... ( -37.10) >DroSim_CAF1 103362 118 - 1 AGCCCACACA--CACGCGCGCCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGGCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACAC .(((((((..--..(((..(((((((.((((((...........)))))))))....((.(....).))...)))).....)))((.......))........))))))).......... ( -34.80) >DroEre_CAF1 102735 118 - 1 AGCCCACACA--CACGCGCGGCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACAC .((((((...--....(((.((.(((.((((((...........))))))))).......(....)(((....))).....)).))).........((....)))))))).......... ( -35.20) >DroYak_CAF1 90835 120 - 1 AGCCCACACACACACGCACGCCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACAC .(((((((......(((.((((.(((.((((((...........)))))))))....((.(....)(((....))).....))))))......))).......))))))).......... ( -37.92) >consensus AGCCCACACA__CACGCGCGCCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAAAGCGGCGGAGAAAGCGGAGAAAUUGUGGGUGAGAAAACAC .(((((((......(((.(.((.((((((((((...........))))))..........(....)(((....))).....)))).)).)...))).......))))))).......... (-32.76 = -33.56 + 0.80)


| Location | 3,712,612 – 3,712,719 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -42.83 |
| Consensus MFE | -37.75 |
| Energy contribution | -38.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3712612 107 + 23771897 UUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGGCGCGCGUGUGUGUGGGCUGGAAAAGUGGGCGUGGCAAAUUGGAAGCA .....(((....((..((((((((((((..(.(.(((..........((((((((......))).))))).)))).)..))))))))))))..))....)))..... ( -39.70) >DroSec_CAF1 107678 107 + 1 UUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGGCGCGCGUGUGUGUGGGCUGGAAAAGUGGGCGUGGCAAUGUGGGAGCA ...(.((((...((..((((((((((((..(.(.(((..........((((((((......))).))))).)))).)..))))))))))))..))...)))).)... ( -41.70) >DroSim_CAF1 103402 107 + 1 UUAUGCCAUUUGCCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGGCGCGCGUGUGUGUGGGCUGGAAAAGUGGGCGUGGCAAUGUGGGAGCA ...(.(((((((((((.(((((((((((..(.(.(((..........((((((((......))).))))).)))).)..)))))))))))))))))).)))).)... ( -43.70) >DroEre_CAF1 102775 106 + 1 UUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGCCGCGCGUGUGUGUGGGCUGGAAAAGUGGGCGUGGCCAUAUAGGCGA- ...((((((.((((..((((((((((((..(.(.(((..........((((((((......))).))))).)))).)..))))))))))))..)))).)).)))).- ( -46.20) >consensus UUAUGCCAUUUGGCGUUGCCUACUUUUCUGGGCGCAUCUAUAAAAUUUACACCGCAGUGGCGCGCGUGUGUGUGGGCUGGAAAAGUGGGCGUGGCAAUGUGGGAGCA ...((((((........(((((((((((..(.(.(((..........((((((((......))).))))).)))).)..)))))))))))))))))........... (-37.75 = -38.00 + 0.25)


| Location | 3,712,612 – 3,712,719 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.35 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3712612 107 - 23771897 UGCUUCCAAUUUGCCACGCCCACUUUUCCAGCCCACACACACGCGCGCCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAA .................(((.(((((((..((.(..((((.((((......))))))))..(((.....)))).))...))))))).)))...(((....))).... ( -26.00) >DroSec_CAF1 107678 107 - 1 UGCUCCCACAUUGCCACGCCCACUUUUCCAGCCCACACACACGCGCGCCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAA .................(((.(((((((..((.(..((((.((((......))))))))..(((.....)))).))...))))))).)))...(((....))).... ( -26.00) >DroSim_CAF1 103402 107 - 1 UGCUCCCACAUUGCCACGCCCACUUUUCCAGCCCACACACACGCGCGCCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGGCAAAUGGCAUAA .....(((..(((((..(((.(((((((..((.(..((((.((((......))))))))..(((.....)))).))...))))))).)))...))))).)))..... ( -29.00) >DroEre_CAF1 102775 106 - 1 -UCGCCUAUAUGGCCACGCCCACUUUUCCAGCCCACACACACGCGCGGCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAA -..(((.((.((((...(((.(((((((..((((.(......).).)))..((.((((((...........))))))))))))))).)))...)))).))))).... ( -35.60) >consensus UGCUCCCACAUUGCCACGCCCACUUUUCCAGCCCACACACACGCGCGCCACUGCGGUGUAAAUUUUAUAGAUGCGCCCAGAAAAGUAGGCAACGCCAAAUGGCAUAA .................(((.(((((((..((.(....((((.((((....))))))))..(((.....)))).))...))))))).)))...(((....))).... (-24.10 = -24.35 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:52 2006