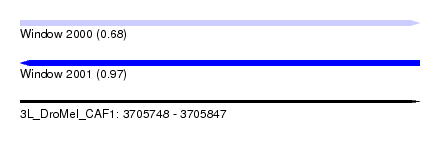
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,705,748 – 3,705,847 |
| Length | 99 |
| Max. P | 0.971498 |

| Location | 3,705,748 – 3,705,847 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -40.28 |
| Consensus MFE | -21.86 |
| Energy contribution | -20.70 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
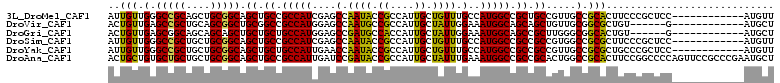
>3L_DroMel_CAF1 3705748 99 + 23771897 AUUGUUGGGCCGCAGCUGCGGCAGCUGCCGCCAUCGAGCCAAUACCGCCAUUGCUGUUUGCCAUGGCCGCUGCCGUUGCCGCACUUCCCGCUCC------------AUGUU ......(((..((.((.((((((((.((((....)).)).......(((((.((.....)).))))).)))))))).)).))....))).....------------..... ( -37.40) >DroVir_CAF1 124604 93 + 1 ACUGUUGAGCCGCUGCAGCGGCUGCGGCCGCCAUGGAGCCAAUGCCGCCAUUGCUAUUGGAAAUGGCAGCAGCUGUUGCGGCGCUGU------G------------AUGCU .......(((.((((((((((((((.(((.(((...(((.((((....)))))))..)))....))).)))))))))))))))))..------.------------..... ( -45.30) >DroGri_CAF1 83416 93 + 1 ACUGUUGAGCGGCAGCAGCAGCUGCUGCUGCCAUGGAGCCGAUGCCACCAUUGCUAUUGGAAAUGGCAGCCGCUUGGGCGGCACUGU------G------------AUGCU .......((((((((((((....)))))))))((((.(((..((((((((.......)))...)))))(((.....)))))).))))------.------------..))) ( -43.00) >DroSim_CAF1 96584 99 + 1 AUUGUUGGGCCGCUGCUGCGGCAGCUGCCGCCAUCGAGCCAAUACCGCCAUUGCUGUUUGCCAUGGCCGCCGCCGUGGCCGCGCUUCCCGCUCC------------AUGUU .......(((.(((((....))))).)))..(((.((((.......(((((.((.....)).)))))(((.((....)).)))......)))).------------))).. ( -39.10) >DroYak_CAF1 83752 99 + 1 AUUGUUGGGCCGCUGCUGCGGCAGCUGCUGCCAUUGAACCAAUACCGCCAUUGCUGUUUGCCAUGGCCGCCGCCGUUGCCGCGCUGCCCGCUCC------------AUGUU ......((((.((....((((((((.((.((.((((...))))...(((((.((.....)).))))).)).)).)))))))))).)))).....------------..... ( -39.90) >DroAna_CAF1 101112 111 + 1 ACUGCUGUGCUGCUGCUGCGGCAGCUGCCGCCAUUGAUCCGAUACCGCCAUUGCUAUUUGAAAUGGCCGCCGCACUGGCCGCACUUCCGGCCCCAGUUCCGCCCGAAUGCU ...((.((((.(((..((((((.((....))...............((((((.(.....).)))))).))))))..))).))))(((.(((.........))).))).)). ( -37.00) >consensus ACUGUUGGGCCGCUGCUGCGGCAGCUGCCGCCAUCGAGCCAAUACCGCCAUUGCUAUUUGAAAUGGCCGCCGCCGUGGCCGCACUGCCCGCUCC____________AUGCU ..(((.(.(((((....))))).((.((.(((((....(.((((.((....)).)))).)..))))).)).)).....).)))............................ (-21.86 = -20.70 + -1.16)
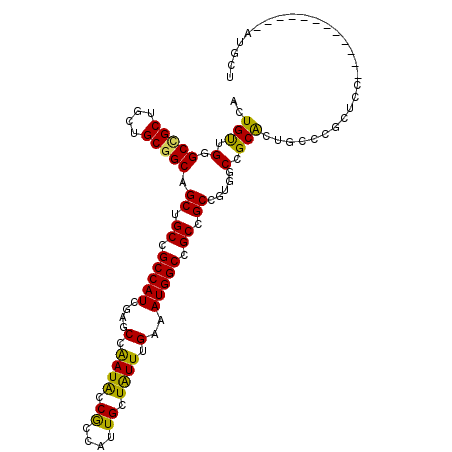

| Location | 3,705,748 – 3,705,847 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -42.47 |
| Consensus MFE | -24.25 |
| Energy contribution | -23.83 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3705748 99 - 23771897 AACAU------------GGAGCGGGAAGUGCGGCAACGGCAGCGGCCAUGGCAAACAGCAAUGGCGGUAUUGGCUCGAUGGCGGCAGCUGCCGCAGCUGCGGCCCAACAAU .....------------.....(((...((((((..(((((((.(((((.((.....)).))))).......((.((....)))).)))))))..)))))).)))...... ( -43.40) >DroVir_CAF1 124604 93 - 1 AGCAU------------C------ACAGCGCCGCAACAGCUGCUGCCAUUUCCAAUAGCAAUGGCGGCAUUGGCUCCAUGGCGGCCGCAGCCGCUGCAGCGGCUCAACAGU .....------------.------...(((((((...(((((((((((((.(.....).)))))))))...)))).....)))).)))((((((....))))))....... ( -40.00) >DroGri_CAF1 83416 93 - 1 AGCAU------------C------ACAGUGCCGCCCAAGCGGCUGCCAUUUCCAAUAGCAAUGGUGGCAUCGGCUCCAUGGCAGCAGCAGCUGCUGCUGCCGCUCAACAGU (((..------------.------...(.(((((....)).((..(((((.(.....).)))))..))...))).)...(((((((((....))))))))))))....... ( -39.70) >DroSim_CAF1 96584 99 - 1 AACAU------------GGAGCGGGAAGCGCGGCCACGGCGGCGGCCAUGGCAAACAGCAAUGGCGGUAUUGGCUCGAUGGCGGCAGCUGCCGCAGCAGCGGCCCAACAAU ..(((------------.((((.((.(.(((.((((.(((....))).))))...((....))))).).)).)))).)))(.(((.(((((....))))).))))...... ( -44.50) >DroYak_CAF1 83752 99 - 1 AACAU------------GGAGCGGGCAGCGCGGCAACGGCGGCGGCCAUGGCAAACAGCAAUGGCGGUAUUGGUUCAAUGGCAGCAGCUGCCGCAGCAGCGGCCCAACAAU .....------------.....((((..(((.((..(((((((.(((((.((.....)).)))))(.(((((...))))).)....)))))))..)).)))))))...... ( -42.40) >DroAna_CAF1 101112 111 - 1 AGCAUUCGGGCGGAACUGGGGCCGGAAGUGCGGCCAGUGCGGCGGCCAUUUCAAAUAGCAAUGGCGGUAUCGGAUCAAUGGCGGCAGCUGCCGCAGCAGCAGCACAGCAGU .((..((((......))))(((((......))))).((((.((.((((((((..(((.(......).)))..))..)))))).)).(((((....))))).)))).))... ( -44.80) >consensus AACAU____________GGAGCGGGAAGCGCGGCAACGGCGGCGGCCAUGGCAAACAGCAAUGGCGGUAUUGGCUCAAUGGCGGCAGCUGCCGCAGCAGCGGCCCAACAAU ...........................(.((.((....((.((.(((((...........))))).))............(((((....))))).)).)).)).)...... (-24.25 = -23.83 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:44 2006