| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 409,239 – 409,350 |
| Length | 111 |
| Max. P | 0.726292 |

| Location | 409,239 – 409,350 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.07 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
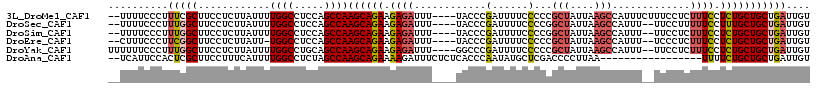
>3L_DroMel_CAF1 409239 111 + 23771897 ACAAUCAGCAGCAGAGGAAAGAGGAAAGAAAUGGCUUAAUAGCGGGGGAAAAUCGGGUA----AAAUCUCUUCUGCUUGGCUGGAGGCCAAAAUAAGAGGAAGCGAAAGGGAAAA-- .......((.((((((((...(((..(....)..)))..((.(.((......)).).))----.....))))))))(((((.....)))))...........))...........-- ( -23.60) >DroSec_CAF1 56138 109 + 1 ACAAUCAGCAGCAAAGGAAAAAGGAA--AAAUGGCUUAAUAGCCGGGGAAAAUCGGGUA----AAAUCUCUUCUGCUUGGCUGGAGGCCAAAAUAAGAGGAAGCCAAAGGGAAAA-- ..........................--...((((((..((.((((......)))).))----...((((((.(..(((((.....))))).).)))))))))))).........-- ( -25.70) >DroSim_CAF1 58252 109 + 1 ACAAUCAGCAGCAGAGGAAAGAGGAA--AAAUGGCUUAAUAGCCGGGGAAAAUCGGGUA----AAAUCUCUUCUGCUUGGCUGGAGGCCAAAAUAAGAGGAAGCCAAAGGGAAAA-- ....((((((((((((((..((....--...(((((....))))).......))((...----...)))))))))))..))))).(((..............)))..........-- ( -28.28) >DroEre_CAF1 60533 108 + 1 ACAAUCAGCAGCAGAGGAAAGAGGGA--AAAUGGCUUAAUAGCGGGGGAAAAUCGGGUA----AAAUCUCUUCUGCUUGGCUGGAGGCCA-AAUAAGAGGAAGCCGAAGGGAAAG-- ....((((((((((((((..((....--...(.(((....))).).......))((...----...)))))))))))..)))))...((.-.......))...((....))....-- ( -25.44) >DroYak_CAF1 60514 111 + 1 ACAAUCAGCAGCAGAGGAAAGAGGAA--AAAUGGCUUAAUAGCGGGGGAAAAUCGGGCC----AAAUCUCUUCUGCUUGGCUGCAGGCCAAAAUAAGAGGAAGCCAAAGGGAAAAAA ....((.(((((.(((...((((((.--...(((((..((..(....)...))..))))----)....)))))).))).))))).(((..............))).....))..... ( -28.24) >DroAna_CAF1 50350 98 + 1 ACAAUCAGCAGCAGAAAA-----------------UUAAGGGGUCGAGCAUAUUGGGUGAGAGAAAUCUUUUCUGCUUGGCUAGAGGCCAAAAUGAAAGGAAGCGAGUGGAAUGA-- .((.((.((.(((((((.-----------------...(((..((...(((.....)))...))..))))))))))(((((.....)))))...........)))).))......-- ( -23.20) >consensus ACAAUCAGCAGCAGAGGAAAGAGGAA__AAAUGGCUUAAUAGCGGGGGAAAAUCGGGUA____AAAUCUCUUCUGCUUGGCUGGAGGCCAAAAUAAGAGGAAGCCAAAGGGAAAA__ .......(((((((((((...............(((....)))..((.....))..............)))))))))((((.....))))............))............. (-18.40 = -18.07 + -0.33)

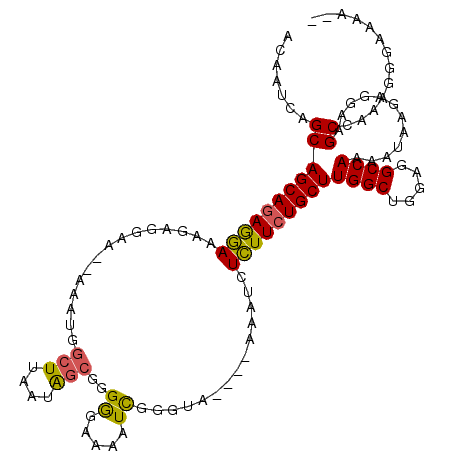
| Location | 409,239 – 409,350 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -14.25 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 409239 111 - 23771897 --UUUUCCCUUUCGCUUCCUCUUAUUUUGGCCUCCAGCCAAGCAGAAGAGAUUU----UACCCGAUUUUCCCCCGCUAUUAAGCCAUUUCUUUCCUCUUUCCUCUGCUGCUGAUUGU --......(..((((............((((.....))))((((((((((....----.....(.....)....(((....)))..........))))....)))))))).))..). ( -17.00) >DroSec_CAF1 56138 109 - 1 --UUUUCCCUUUGGCUUCCUCUUAUUUUGGCCUCCAGCCAAGCAGAAGAGAUUU----UACCCGAUUUUCCCCGGCUAUUAAGCCAUUU--UUCCUUUUUCCUUUGCUGCUGAUUGU --..........((((............))))..((((..((((((.((((...----.....(........)((((....))))....--......)))).))))))))))..... ( -21.80) >DroSim_CAF1 58252 109 - 1 --UUUUCCCUUUGGCUUCCUCUUAUUUUGGCCUCCAGCCAAGCAGAAGAGAUUU----UACCCGAUUUUCCCCGGCUAUUAAGCCAUUU--UUCCUCUUUCCUCUGCUGCUGAUUGU --..........((((............))))..((((..((((((((((....----.....(........)((((....))))....--...))))....))))))))))..... ( -24.30) >DroEre_CAF1 60533 108 - 1 --CUUUCCCUUCGGCUUCCUCUUAUU-UGGCCUCCAGCCAAGCAGAAGAGAUUU----UACCCGAUUUUCCCCCGCUAUUAAGCCAUUU--UCCCUCUUUCCUCUGCUGCUGAUUGU --........(((((...........-((((.....))))((((((((((....----.....(.....)....(((....))).....--...))))....))))))))))).... ( -21.40) >DroYak_CAF1 60514 111 - 1 UUUUUUCCCUUUGGCUUCCUCUUAUUUUGGCCUGCAGCCAAGCAGAAGAGAUUU----GGCCCGAUUUUCCCCCGCUAUUAAGCCAUUU--UUCCUCUUUCCUCUGCUGCUGAUUGU ............((((............)))).(((((..((.(((.((((..(----(((..(((...........)))..))))..)--))).)))...))..)))))....... ( -24.00) >DroAna_CAF1 50350 98 - 1 --UCAUUCCACUCGCUUCCUUUCAUUUUGGCCUCUAGCCAAGCAGAAAAGAUUUCUCUCACCCAAUAUGCUCGACCCCUUAA-----------------UUUUCUGCUGCUGAUUGU --......((.((((............((((.....))))(((((((((........((.............))........-----------------))))))))))).)).)). ( -16.11) >consensus __UUUUCCCUUUGGCUUCCUCUUAUUUUGGCCUCCAGCCAAGCAGAAGAGAUUU____UACCCGAUUUUCCCCCGCUAUUAAGCCAUUU__UUCCUCUUUCCUCUGCUGCUGAUUGU ..........(((((............((((.....))))((((((.((((............(......)...(((....))).............)))).))))))))))).... (-14.25 = -14.75 + 0.50)
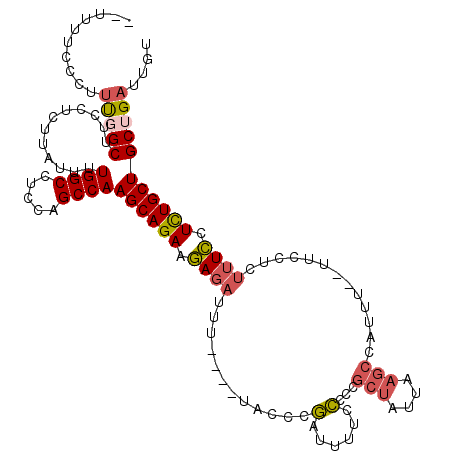
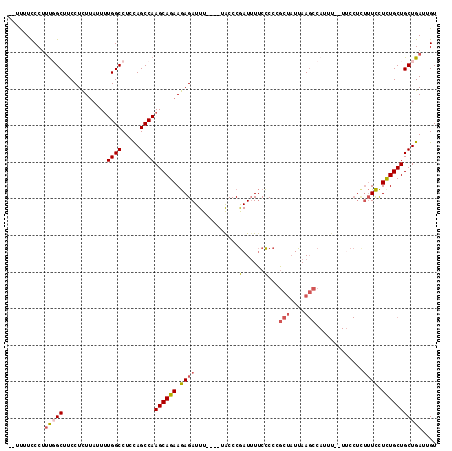
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:18 2006