
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,689,165 – 3,689,300 |
| Length | 135 |
| Max. P | 0.921849 |

| Location | 3,689,165 – 3,689,265 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -15.59 |
| Energy contribution | -17.60 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
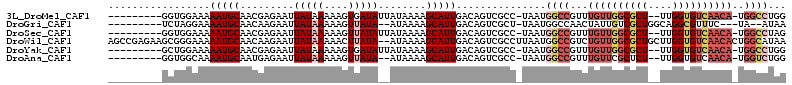
>3L_DroMel_CAF1 3689165 100 + 23771897 ---------GGUGGAAAAAUGCAACGAGAAUUAUAAAAAGUGAUAUUAUAAAAGCAUUGACAGUCGCC-UAAUGGCCGUUUGUUGGCGCU--UUGGUGUCAACA-UGGCCUGG ---------(((((...(((((....((.(((((.....))))).))......))))).....)))))-....(((((..(((((((((.--...)))))))))-)))))... ( -30.60) >DroGri_CAF1 65784 96 + 1 ---------UCUAGGAAAAUGCAACAAGAAUUAUAAAAAGUUAUA--AUAAAAGCAUUGACAGUCGCU-UAAUGGCCAACUAUUGUCGCUGGCAGGCGUUUC---UA--AUAA ---------..((((((..(((.......(((((((....)))))--)).....((.(((((((.(((-....))).....))))))).)))))....))))---))--.... ( -17.70) >DroSec_CAF1 84543 100 + 1 ---------GGUGGAAAAAUGCAACGAGAAUUAUAAAAAGUUAUAUUAUAAAAGCAUUGACAGUCGCC-UAAUGGCCGUUUGUUGGCGCU--UUGGUGUCAACA-UGGCCUAG ---------(((((...(((((.........(((((....)))))........))))).....)))))-....(((((..(((((((((.--...)))))))))-)))))... ( -30.83) >DroWil_CAF1 111277 111 + 1 AGCCGAGAAGCGGGAAAAAUGCAACAAGAAUUAUAAAAACUUAUA--AUAAAAGCAUUGACAGUCGCCUUAAUGGCCGUCUGUUGGCGCUGCUUGGUGUCAACACUGGCAUAA .((((((.((((.....(((((.......(((((((....)))))--))....)))))(((((.((((.....)).)).)))))..)))).))))))((((....)))).... ( -32.70) >DroYak_CAF1 66625 100 + 1 ---------GCUGGAAAAAUGCAACGAGAAUUAUAAAAAGUGAUAUUAUAAAAGCAUUGACAGUCGCC-UAAUGGCCGUUUGUUGGCGCU--UUGGUGUCAACA-UGGCCUGG ---------((((....(((((....((.(((((.....))))).))......)))))..))))..((-....(((((..(((((((((.--...)))))))))-))))).)) ( -28.40) >DroAna_CAF1 83058 98 + 1 ---------GGUGGCAAAAUGCAAUGAGAAUUAUAAAAAGUUAUA--AUAAAAGCAUUGACAGUCGCC-UAAUGGCCGUUUGUUCGCUCU--UUGGUGUCAACA-UGGUCUGG ---------((((((....(((((((...(((((((....)))))--)).....))))).))))))))-....(((((((((..((((..--..)))).))).)-)))))... ( -25.20) >consensus _________GCUGGAAAAAUGCAACGAGAAUUAUAAAAAGUUAUA__AUAAAAGCAUUGACAGUCGCC_UAAUGGCCGUUUGUUGGCGCU__UUGGUGUCAACA_UGGCCUAG .................(((((.........(((((....)))))........)))))...............((((...((((((((((....))))))))))..))))... (-15.59 = -17.60 + 2.00)

| Location | 3,689,196 – 3,689,300 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.15 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -26.59 |
| Energy contribution | -27.28 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
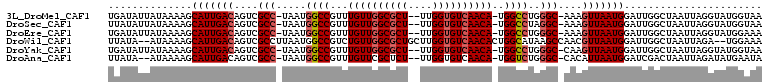
>3L_DroMel_CAF1 3689196 104 + 23771897 UGAUAUUAUAAAAGCAUUGACAGUCGCC-UAAUGGCCGUUUGUUGGCGCU--UUGGUGUCAACA-UGGCCUGGGC-AAAGUUAAUGGAUUGGCUAAUUAGGUAUGGUAA ...((((((.....(((((((..(.(((-((..(((((..(((((((((.--...)))))))))-))))))))))-)..))))))).....(((.....))))))))). ( -34.90) >DroSec_CAF1 84574 104 + 1 UUAUAUUAUAAAAGCAUUGACAGUCGCC-UAAUGGCCGUUUGUUGGCGCU--UUGGUGUCAACA-UGGCCUAGGC-AAAGUUAAUGGAUUGGCUAAUUAGGUAUGGUAA ...((((((.....(((((((..(.(((-((..(((((..(((((((((.--...)))))))))-))))))))))-)..))))))).....(((.....))))))))). ( -35.20) >DroEre_CAF1 79138 104 + 1 UGAUAUUAUAAAAGCAUUGACAGUCGCC-UAAUGGCCGUUUGUUGGCGCU--UUGGUGUCAACA-UGGCCUGGGC-AAAGUUAAUGGAUUGGCUAAUUAGGUAUGGAAA ..............(((((((..(.(((-((..(((((..(((((((((.--...)))))))))-))))))))))-)..))))))).....(((.....)))....... ( -33.20) >DroWil_CAF1 111317 105 + 1 UUAUA--AUAAAAGCAUUGACAGUCGCCUUAAUGGCCGUCUGUUGGCGCUGCUUGGUGUCAACACUGGCAUAAGCCAACGUUAAUGGAUUGGCUAAUUAGA--UGGAAA .....--.......(((((((((.((((.....)).)).))((((((..((((.((((....))))))))...)))))))))))))...............--...... ( -30.40) >DroYak_CAF1 66656 104 + 1 UGAUAUUAUAAAAGCAUUGACAGUCGCC-UAAUGGCCGUUUGUUGGCGCU--UUGGUGUCAACA-UGGCCUGGGC-CAAGUUAAUGGAUUGGCUAAUUAGGUAUGGUAA ....................((...(((-(((((((((..(((((((((.--...)))))))))-)))))..(((-(((.........)))))).))))))).)).... ( -35.30) >DroAna_CAF1 83089 102 + 1 UUAUA--AUAAAAGCAUUGACAGUCGCC-UAAUGGCCGUUUGUUCGCUCU--UUGGUGUCAACA-UGGUCUGGGC-CACAUUAAUGGAUCGACUAAUUAGAUAUGAAUA .....--.......((((((..((.(((-((..(((((((((..((((..--..)))).))).)-))))))))))-.)).))))))....................... ( -22.00) >consensus UGAUAUUAUAAAAGCAUUGACAGUCGCC_UAAUGGCCGUUUGUUGGCGCU__UUGGUGUCAACA_UGGCCUGGGC_AAAGUUAAUGGAUUGGCUAAUUAGGUAUGGAAA ..............(((((((....(((.....((((...((((((((((....))))))))))..))))..)))....)))))))....................... (-26.59 = -27.28 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:38 2006