| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,647,035 – 3,647,125 |
| Length | 90 |
| Max. P | 0.874948 |

| Location | 3,647,035 – 3,647,125 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.22 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -11.35 |
| Energy contribution | -11.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
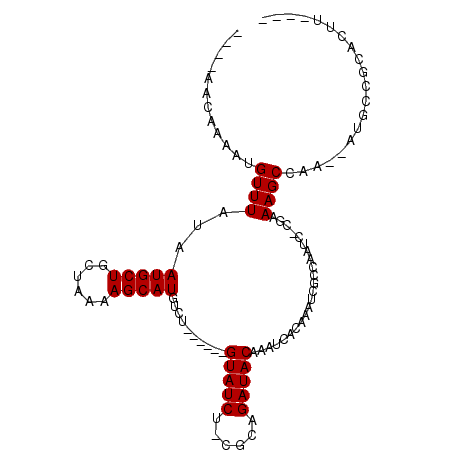
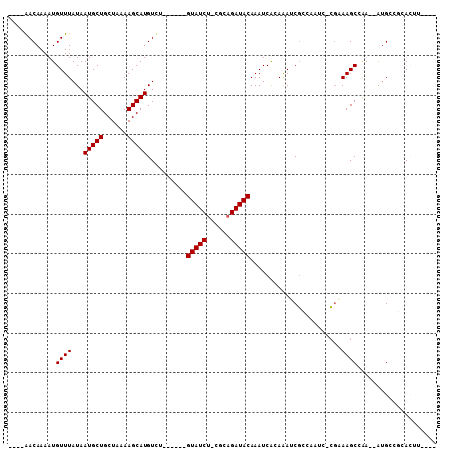
>3L_DroMel_CAF1 3647035 90 + 23771897 ----AACAAAAUGUUUAUAAUGCUGCUAAAAGCAUGUUU------GUAUCU-CGCAGAUACAAAUCGCAGAUCGCCAAUC-CGAAAGCCAA--AUGCCGCACUU---- ----................(((.((.....((..((((------(((((.-....))))))))).))...(((......-))).......--..)).)))...---- ( -16.80) >DroVir_CAF1 44094 97 + 1 GCGAGACAAAAUGUUUAUAAUGCUGCUAAAAGCAUGUCAGUACGAGUAUCU-CGCAGAUACAAAUCAGAAAGCGGC---C-CGCAAGCAAA--GUGGGCCACGA---- ((((((.(...(((.....(((((......)))))......)))..).)))-))).(((....))).....(.(((---(-(((.......--))))))).)..---- ( -31.10) >DroGri_CAF1 18264 98 + 1 GCGAGACAAAAUGUUUAUAAUGCUGCUAAAAGCAUGUCAGUACGAGUAUCU-CGCAGAUACAAAUCAGAAAGCGGC---CGCUCAAGCAAA--GUGGGCCACGA---- ((((((.(...(((.....(((((......)))))......)))..).)))-))).(((....))).....(.(((---((((.......)--)).)))).)..---- ( -24.80) >DroWil_CAF1 48507 94 + 1 ----AACAAAAUGUUUAUAAUGCUGCUAAAAGCAUGUCC------GUAUCGGUCCAGAUACAAAUCAGAAAACUGAAAAC-AGAAAGCCGA--AAGUGG-AUUCUGUC ----...............(((((......)))))....------(((((......)))))...((((....))))..((-((((..((..--..).).-.)))))). ( -20.60) >DroYak_CAF1 22396 90 + 1 ----AACAAAAUGUUUAUAAUGCUGCUAAAAGCAUGUUU------GUAUCU-CGCAGAUACAAAUCGCAGAUCGCCAAUC-CGCAAGCCAA--AUGCCGCACUU---- ----................(((.((.....((..((((------(((((.-....))))))))).((.(((.....)))-.))..))...--..)).)))...---- ( -19.00) >DroAna_CAF1 39320 92 + 1 ----AACAAAAUGUUUAUAAUGCUGCUAAAAGCAUGUUU------GUAUCU-UGCAGAUACAAAUCGCAGAUCGCAAAUC-CGAAAGCCAAAAAUGCCGAAACU---- ----........((((...(((((......)))))((((------(((((.-....))))))))).((((((.....)))-(....).......)))..)))).---- ( -16.20) >consensus ____AACAAAAUGUUUAUAAUGCUGCUAAAAGCAUGUCU______GUAUCU_CGCAGAUACAAAUCACAAAUCGCCAAUC_CGAAAGCCAA__AUGCCGCACUU____ ............((((...(((((......)))))..........(((((......))))).......................)))).................... (-11.35 = -11.35 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:17 2006