| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
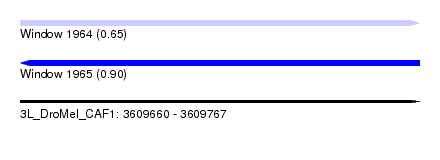
| Location | 3,609,660 – 3,609,767 |
| Length | 107 |
| Max. P | 0.901071 |

| Location | 3,609,660 – 3,609,767 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -36.22 |
| Consensus MFE | -27.60 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
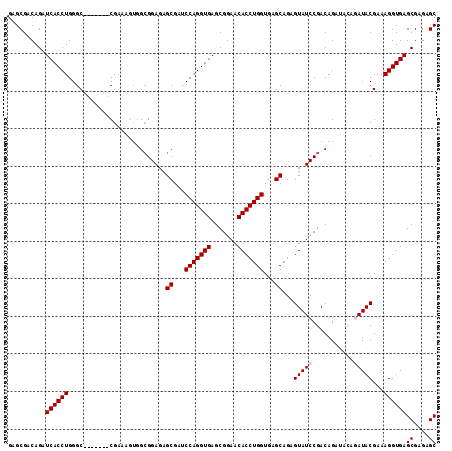
>3L_DroMel_CAF1 3609660 107 + 23771897 GAGCGACAGAUCACCUGGGC-------CGAAAGUGGCGGAGAGCGGACCAGGUGAGCGGAACACCUGGUGAGCAGAGUAUCCGACAGAUACAGAUACGAAAGGUGAGCGAGAGC ..........((((((..((-------((....)))).....((..((((((((.......))))))))..))...(((((.....))))).........))))))((....)) ( -36.60) >DroSec_CAF1 134263 107 + 1 GAGCGACAGAUCACCUGGGC-------CGAAAGUGGCGGAGAGCGAUCCAGGUGAGCGGAACACCUGGUGAGCAGAGUAUCCGACAGAUACAGAUACGAAAGGUGAGCGAGAGC ..........((((((..((-------((....)))).....((...(((((((.......)))))))...))...(((((.....))))).........))))))((....)) ( -34.60) >DroSim_CAF1 140166 107 + 1 GAGCGACAGAUCACCUGGGC-------CGAAAGUCGCGGAGAGCGAUCCAGGUGAGCGGAACACCUGGUGAGCAGAGUAUCCGACAGAUACAGAUACGAAAGGUGAGCGAGAGC ..((..(...(((((((((.-------(....)((((.....)))))))))))))...)..(((((..((......(((((.....))))).....))..))))).))...... ( -35.50) >DroEre_CAF1 156630 101 + 1 GAGCGUCAGAUCACCUGGGC-------CGAAACUGGCGGAGAGCGAUCCAGGUGAGCGGAACACCUGGUGAGCAGAGUAUCCGACAG------AUACGAAAGGUGAGCGAGAGC ..........((((((..((-------((....)))).....((...(((((((.......)))))))...))...(((((.....)------))))...))))))((....)) ( -34.80) >DroYak_CAF1 139203 108 + 1 GAGCGACAGAUCACCUGGGCCUGGGCCCGAAACGCGAGGAGAGCGAUCCAGGUGAGCGGAACACCUGGUGAGCAGAGUAUCCGACAG------AUACGAAAGGUGAGCGAGAGC ..........((((((((((....))))....(....)....((...(((((((.......)))))))...))...(((((.....)------))))...))))))((....)) ( -39.60) >consensus GAGCGACAGAUCACCUGGGC_______CGAAAGUGGCGGAGAGCGAUCCAGGUGAGCGGAACACCUGGUGAGCAGAGUAUCCGACAGAUACAGAUACGAAAGGUGAGCGAGAGC ..........((((((..........................((...(((((((.......)))))))...))...(((((...........)))))...))))))((....)) (-27.60 = -28.00 + 0.40)

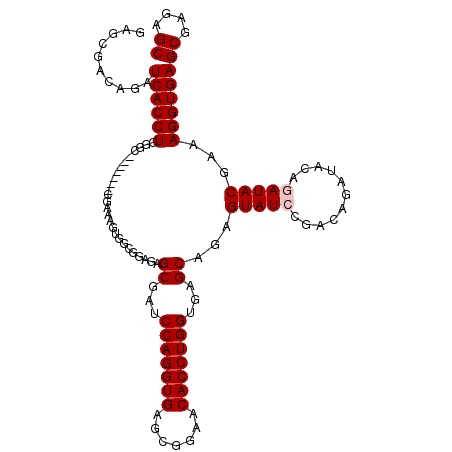
| Location | 3,609,660 – 3,609,767 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -25.34 |
| Energy contribution | -26.54 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3609660 107 - 23771897 GCUCUCGCUCACCUUUCGUAUCUGUAUCUGUCGGAUACUCUGCUCACCAGGUGUUCCGCUCACCUGGUCCGCUCUCCGCCACUUUCG-------GCCCAGGUGAUCUGUCGCUC ((....(.((((((...(((((((.......)))))))...((..((((((((.......))))))))..)).....(((......)-------))..)))))).)....)).. ( -35.10) >DroSec_CAF1 134263 107 - 1 GCUCUCGCUCACCUUUCGUAUCUGUAUCUGUCGGAUACUCUGCUCACCAGGUGUUCCGCUCACCUGGAUCGCUCUCCGCCACUUUCG-------GCCCAGGUGAUCUGUCGCUC ((....(.((((((...(((((((.......)))))))...((...(((((((.......)))))))...)).....(((......)-------))..)))))).)....)).. ( -33.00) >DroSim_CAF1 140166 107 - 1 GCUCUCGCUCACCUUUCGUAUCUGUAUCUGUCGGAUACUCUGCUCACCAGGUGUUCCGCUCACCUGGAUCGCUCUCCGCGACUUUCG-------GCCCAGGUGAUCUGUCGCUC ((....(.((((((...(((((((.......)))))))...(((..(((((((.......))))))).((((.....)))).....)-------))..)))))).)....)).. ( -34.40) >DroEre_CAF1 156630 101 - 1 GCUCUCGCUCACCUUUCGUAU------CUGUCGGAUACUCUGCUCACCAGGUGUUCCGCUCACCUGGAUCGCUCUCCGCCAGUUUCG-------GCCCAGGUGAUCUGACGCUC ((((....((((((.(((...------(((.((((......((...(((((((.......)))))))...))..)))).)))...))-------)...))))))...)).)).. ( -33.60) >DroYak_CAF1 139203 108 - 1 GCUCUCGCUCACCUUUCGUAU------CUGUCGGAUACUCUGCUCACCAGGUGUUCCGCUCACCUGGAUCGCUCUCCUCGCGUUUCGGGCCCAGGCCCAGGUGAUCUGUCGCUC ((....(.((((((...((((------(.....)))))........(((((((.......)))))))..(((.......)))....((((....)))))))))).)....)).. ( -36.10) >consensus GCUCUCGCUCACCUUUCGUAUCUGUAUCUGUCGGAUACUCUGCUCACCAGGUGUUCCGCUCACCUGGAUCGCUCUCCGCCACUUUCG_______GCCCAGGUGAUCUGUCGCUC ((....(.((((((...(((((((.......)))))))...((...(((((((.......)))))))...))..........................)))))).)....)).. (-25.34 = -26.54 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:10 2006