
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
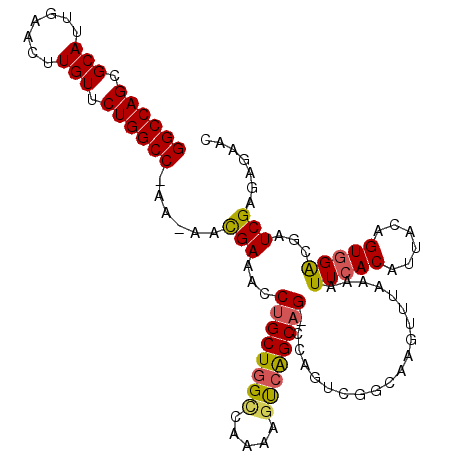
| Location | 3,596,444 – 3,596,575 |
| Length | 131 |
| Max. P | 0.983955 |

| Location | 3,596,444 – 3,596,550 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 90.22 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -27.41 |
| Energy contribution | -27.22 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3596444 106 - 23771897 GGCCAGCGCAUUGAACUUGUUCUGGCC-AA-AACGAAACCUGCUGGCCAAAAGUCAGCAG-CCAGUCGGCAAGUUUAAAAUUCACAUUACAGUGGGCGAUCGAGAGAAC .(((......((((((((((.(((((.-..-.........(((((((.....))))))))-))))...))))))))))....(((......))))))..((....)).. ( -35.10) >DroSec_CAF1 121022 106 - 1 GGCCAGCGCAUUGAACUUGUUCUGGCC-AA-AAUGAAACCUGCUGGCCAAAAGUCAGCAG-CCAGUCGGCAAGUUUUAAAUUCACAUUACAGUGGGCGAUCGAGAGAAC ((((((.(((.......))).))))))-..-..((((((.(((((((.....)))))))(-((....)))..))))))..(((((......)))))...((....)).. ( -36.20) >DroSim_CAF1 126878 106 - 1 GGCCAGCGCAUUGAACUUGUUCUGGCC-AA-AACGAAACCUGCUGGCCAAAAGUCAGCAG-CCAGUCGGCAAGUUUAAAAUUCACAUUACAGUGGGCGAUCGAGAGAAC .(((......((((((((((.(((((.-..-.........(((((((.....))))))))-))))...))))))))))....(((......))))))..((....)).. ( -35.10) >DroEre_CAF1 142421 106 - 1 GGCCAGCGCAUUGAACUUGUUCUGGCC-AA-AACGAAACCUGCUGGCCAAAAGCGGGCAG-CCAGUCAGUAAGACUUAAAUUCACAUUACAGUGGACGAUCGAGAGAGC ((((((.(((.......))).))))))-..-..((....(((((.((.....)).)))))-..((((.....))))....(((((......))))))).((....)).. ( -33.00) >DroYak_CAF1 125483 107 - 1 GGCCAGCGCAUUGAACUUGUUCUGGCC-AAAAACGAAACCUGCUGGUUAAAAGUCAGCAG-CCAGUCGCCAAGUUUAAAAUUCACAAUACAGUGGACGAUCGAGAGAGC ((((((.(((.......))).))))))-.....(((...(((((((.......)))))))-....)))............(((((......)))))...((....)).. ( -30.90) >DroAna_CAF1 135083 108 - 1 GGCCAGCGCAUUGAACUUGUUCUGGCCCAA-AACGAAACCUGCUGGCCAAAACCCAGCUGGCGCGUCACCCAGCUUAAAAUUCACAAUACAGUAGACGAUCGGCAGAGC ((((((.(((.......))).))))))...-........((((((..(.......((((((........))))))......((((......)).)).)..))))))... ( -32.40) >consensus GGCCAGCGCAUUGAACUUGUUCUGGCC_AA_AACGAAACCUGCUGGCCAAAAGUCAGCAG_CCAGUCGGCAAGUUUAAAAUUCACAUUACAGUGGACGAUCGAGAGAAC ((((((.(((.......))).))))))......(((...((((((((.....))))))))....................(((((......)))))...)))....... (-27.41 = -27.22 + -0.19)


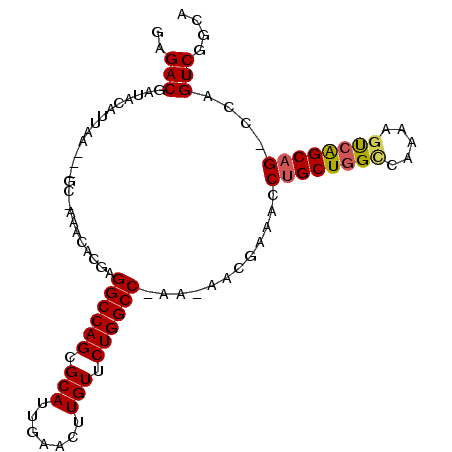
| Location | 3,596,482 – 3,596,575 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.63 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3596482 93 - 23771897 GAGACGCUACAUUAA---GC-AAACACGAGGCCAGCGCAUUGAACUUGUUCUGGCC-AA-AACGAAACCUGCUGGCCAAAAGUCAGCAG-CCAGUCGGCA .....(((......)---))-........((((((.(((.......))).))))))-..-..(((...((((((((.....))))))))-....)))... ( -32.90) >DroSec_CAF1 121060 93 - 1 GAGACGCGACAUUAA---GC-AAAUACGAGGCCAGCGCAUUGAACUUGUUCUGGCC-AA-AAUGAAACCUGCUGGCCAAAAGUCAGCAG-CCAGUCGGCA .....(((((.....---..-........((((((.(((.......))).))))))-..-........((((((((.....))))))))-...))).)). ( -32.60) >DroSim_CAF1 126916 93 - 1 GAGACGCGACAUUAA---GC-AAACACGAGGCCAGCGCAUUGAACUUGUUCUGGCC-AA-AACGAAACCUGCUGGCCAAAAGUCAGCAG-CCAGUCGGCA .....((........---..-........((((((.(((.......))).))))))-..-..(((...((((((((.....))))))))-....))))). ( -32.80) >DroEre_CAF1 142459 93 - 1 GAGACGAUACAUUAA---GC-AAACACGAGGCCAGCGCAUUGAACUUGUUCUGGCC-AA-AACGAAACCUGCUGGCCAAAAGCGGGCAG-CCAGUCAGUA ..(((..........---..-........((((((.(((.......))).))))))-..-........(((((.((.....)).)))))-...))).... ( -28.80) >DroYak_CAF1 125521 95 - 1 AAGACGAUACAUUAA---GCCAAACACGAGGCCAGCGCAUUGAACUUGUUCUGGCC-AAAAACGAAACCUGCUGGUUAAAAGUCAGCAG-CCAGUCGCCA ..(((..........---...........((((((.(((.......))).))))))-...........(((((((.......)))))))-...))).... ( -27.10) >DroAna_CAF1 135121 98 - 1 AAGACGAACGAUUAAAAGGC-AAGCACGAGGCCAGCGCAUUGAACUUGUUCUGGCCCAA-AACGAAACCUGCUGGCCAAAACCCAGCUGGCGCGUCACCC ..((((..............-......(.((((((.(((.......))).)))))))..-.......((.(((((.......))))).))..)))).... ( -29.50) >consensus GAGACGAUACAUUAA___GC_AAACACGAGGCCAGCGCAUUGAACUUGUUCUGGCC_AA_AACGAAACCUGCUGGCCAAAAGUCAGCAG_CCAGUCGGCA ..(((........................((((((.(((.......))).))))))............((((((((.....))))))))....))).... (-26.60 = -26.63 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:09 2006