
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,584,693 – 3,584,788 |
| Length | 95 |
| Max. P | 0.997889 |

| Location | 3,584,693 – 3,584,788 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.59 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -16.00 |
| Energy contribution | -18.12 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3584693 95 + 23771897 AUACUAAAUUCCGUUUUUCCCGAGGA----CCUGCAUUCAUUAGCA-GA-CCUCCU-AUCGGAUG---------------CCACUCGGUAAAUGGAAACAUUUCCGAAAAAAGGAAA ........((((.((((((..((((.----.((((........)))-).-))))..-...(((((---------------((....))))((((....))))))))))))).)))). ( -29.30) >DroPse_CAF1 154942 108 + 1 ACAUUCACUUCCGACAG---CCAGGAUACUCGGGUAUUCAUUAGGAAGAGCCGCCC-AUUCGCUUUCUCUGGGCGAAAGAACAUUUUGUAAAUGGAAACAGUUUCGAGAAA-----G ...(((.((((((....---.).((((((....))))))....)))))...(((((-(...........))))))...)))..(((((....((....))....)))))..-----. ( -27.40) >DroSec_CAF1 114540 95 + 1 AUACUAAAUUCCGUUUUUCCCGAGGA----CCUGCAUUCAUUAGCA-GA-CCGCCU-AUCGGAUG---------------CCACUCGGUAAAUGGAAACAUUUCCGAAAAAAGGAAA ........((((.(((((((((((..----...((((((((..((.-..-..))..-)).)))))---------------)..))))).(((((....)))))..)))))).)))). ( -28.70) >DroSim_CAF1 120392 95 + 1 AUACUAAAUUCCGUUUUUCCCGAGGA----CCUGCAUUUAUUAGCA-GA-CCUCCU-AUCGGAUG---------------CCACUCGGUAAAUGGAAACAUUUCCAAAAAAAGGAAA ........((((((((...(((((..----.((((........)))-).-..(((.-...)))..---------------...))))).))))))))...(((((.......))))) ( -26.00) >DroEre_CAF1 135903 86 + 1 -------CUUCCGUCUUUCCCGAGGA----CCUGCAUUCAUUAGCA-GA-CCUCCU-AUCGGAUG---------------CCACUCGGUAAAUGGAAACAUUUCCGAA--AAGGAUA -------..(((((((.....((((.----.((((........)))-).-))))..-...)))).---------------....((((.(((((....))))))))).--..))).. ( -27.50) >DroYak_CAF1 118905 89 + 1 -------CUUCCGUUUAUCCCGAGGA----CCUGCAUUCAUUAGCA-GA-CCUCCUCAUCGGAUG---------------CCACUCGGUAAAUGGAAACAUUUCCGAAAAAAGGAAA -------.((((...(((((.(((((----.((((........)))-).-..)))))...)))))---------------....((((.(((((....))))))))).....)))). ( -31.80) >consensus AUACUAAAUUCCGUUUUUCCCGAGGA____CCUGCAUUCAUUAGCA_GA_CCUCCU_AUCGGAUG_______________CCACUCGGUAAAUGGAAACAUUUCCGAAAAAAGGAAA ........((((.......(((((((......(((........)))......)))...))))......................((((.(((((....))))))))).....)))). (-16.00 = -18.12 + 2.11)
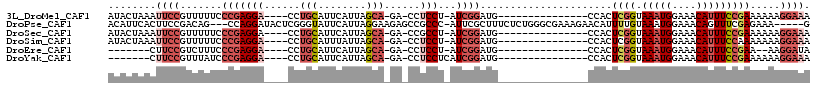
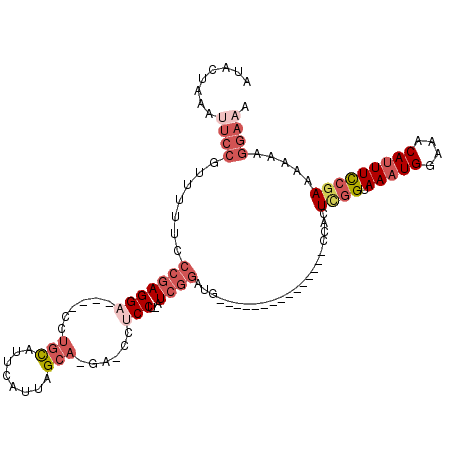

| Location | 3,584,693 – 3,584,788 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.59 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -12.32 |
| Energy contribution | -15.38 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3584693 95 - 23771897 UUUCCUUUUUUCGGAAAUGUUUCCAUUUACCGAGUGG---------------CAUCCGAU-AGGAGG-UC-UGCUAAUGAAUGCAGG----UCCUCGGGAAAAACGGAAUUUAGUAU .((((.((((((((((....)))).....(((((.((---------------(.(((...-.)))..-.(-(((........)))))----))))))))))))).))))........ ( -33.10) >DroPse_CAF1 154942 108 - 1 C-----UUUCUCGAAACUGUUUCCAUUUACAAAAUGUUCUUUCGCCCAGAGAAAGCGAAU-GGGCGGCUCUUCCUAAUGAAUACCCGAGUAUCCUGG---CUGUCGGAAGUGAAUGU .-----.................(((((((....(((((....(((((..(....)...)-))))((.....))....))))).((((((......)---)..))))..))))))). ( -22.00) >DroSec_CAF1 114540 95 - 1 UUUCCUUUUUUCGGAAAUGUUUCCAUUUACCGAGUGG---------------CAUCCGAU-AGGCGG-UC-UGCUAAUGAAUGCAGG----UCCUCGGGAAAAACGGAAUUUAGUAU .((((.((((((((((....)))).....(((((.((---------------(..(((..-...)))-.(-(((........)))))----))))))))))))).))))........ ( -32.30) >DroSim_CAF1 120392 95 - 1 UUUCCUUUUUUUGGAAAUGUUUCCAUUUACCGAGUGG---------------CAUCCGAU-AGGAGG-UC-UGCUAAUAAAUGCAGG----UCCUCGGGAAAAACGGAAUUUAGUAU .((((.((((((((((....)))).....(((((.((---------------(.(((...-.)))..-.(-(((........)))))----))))))))))))).))))........ ( -30.60) >DroEre_CAF1 135903 86 - 1 UAUCCUU--UUCGGAAAUGUUUCCAUUUACCGAGUGG---------------CAUCCGAU-AGGAGG-UC-UGCUAAUGAAUGCAGG----UCCUCGGGAAAGACGGAAG------- ..(((((--(((((((....)))).....(((((.((---------------(.(((...-.)))..-.(-(((........)))))----))))))))))))..)))..------- ( -29.10) >DroYak_CAF1 118905 89 - 1 UUUCCUUUUUUCGGAAAUGUUUCCAUUUACCGAGUGG---------------CAUCCGAUGAGGAGG-UC-UGCUAAUGAAUGCAGG----UCCUCGGGAUAAACGGAAG------- .((((.(((.((((((....)))).....(((((.((---------------(.(((.....)))..-.(-(((........)))))----))))))))).))).)))).------- ( -30.00) >consensus UUUCCUUUUUUCGGAAAUGUUUCCAUUUACCGAGUGG_______________CAUCCGAU_AGGAGG_UC_UGCUAAUGAAUGCAGG____UCCUCGGGAAAAACGGAAGUUAGUAU ..(((.((((((((((....)))).....(((((....................(((.....)))......(((........)))........))))))))))).)))......... (-12.32 = -15.38 + 3.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:07 2006