
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,537,761 – 3,537,928 |
| Length | 167 |
| Max. P | 0.840161 |

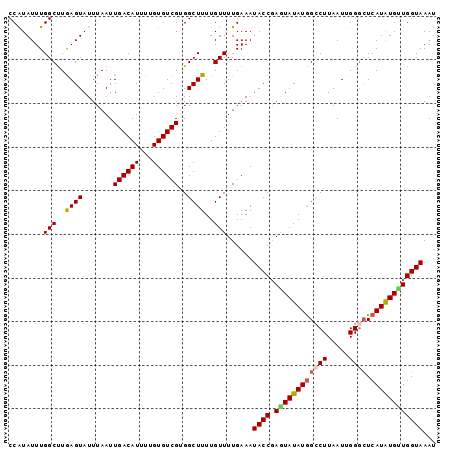
| Location | 3,537,761 – 3,537,859 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -18.66 |
| Energy contribution | -19.06 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3537761 98 + 23771897 CCAUAUUUGGCUUAAGUAUUUAAUUGACAUUUUGUGUCGUGGCUUUUGUUUUGAAAUACCGAGUAUAUGGCCUUAAUUGGGAUCAUGUGUUGGUAAAU ........(((..((((.......((((((...))))))..))))..)))......((((((.((((((((((.....))).)))))))))))))... ( -21.90) >DroSec_CAF1 67926 97 + 1 CCAUAUUUGGCUUGAGUAUUUAAUUGACAUUUUGUGUCGUGGCUUUUGUUUUGAAAUACCGAGUAUAUGGCCUUAAUUGGGCUCAUAUG-UGGUAAAU ((((((.(((((((.((((((....(((((...)))))..(((....)))...)))))))))))....(((((.....))))))).)))-)))..... ( -25.40) >DroSim_CAF1 69057 98 + 1 CCAUAUUUGGCCUGAGUAUUUAAUUGACAUUUUGUGUCGUGGCUUUUGUUUUGAAAUACCGAGUAUAUGGACUUAAUUGGGCUCAUAUGCUGGUAAAU ..(((...((((............((((((...)))))).))))..))).......((((.((((((((..(((....)))..))))))))))))... ( -22.92) >DroEre_CAF1 85960 98 + 1 CCGUAUUUGGCUUGAGUAUUUAAUUGACAUUUUGUGUCGGGGCUUUUGUUUUGAAAUACCCAAUAUAUGGCCUUAAUUGGACUCAUAUGUUGGUAAAU ..(((((((((..((((......(((((((...))))))).))))..)))...))))))(((((((((((((......)).).))))))))))..... ( -26.00) >DroYak_CAF1 70931 98 + 1 CCAUAUUUGGCUUGAGUAUUUAAUUGACAUUUUGUGUCGGGGCUUUUGUUUUGAAAUACCGAAUAUAUAGCCUUAAUUGGGCUCAUAUGUUGGUAAAU ........(((..((((......(((((((...))))))).))))..)))......((((.((((((((((((.....))))).)))))))))))... ( -24.40) >consensus CCAUAUUUGGCUUGAGUAUUUAAUUGACAUUUUGUGUCGUGGCUUUUGUUUUGAAAUACCGAGUAUAUGGCCUUAAUUGGGCUCAUAUGUUGGUAAAU ........(((..((((.......((((((...))))))..))))..)))......((((.((((((((((((.....)))).))))))))))))... (-18.66 = -19.06 + 0.40)


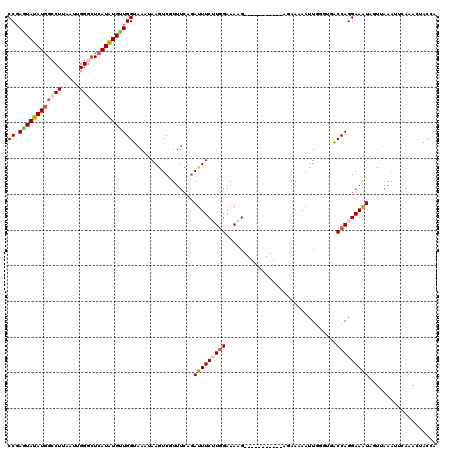
| Location | 3,537,779 – 3,537,888 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3537779 109 + 23771897 UUUAAUUGACAUUUUGUGUCGUGGCUUUUGUUUUGAAAUACCGAGUAUAUGGCCUUAAUUGGGAUCAUGUGUUGGUAAAUAAGUCGUUUCAAAUUUCUUGGAAAAG-----------AGA .......(((........(((..((....))..)))..((((((.((((((((((.....))).))))))))))))).....)))........((((((....)))-----------))) ( -25.60) >DroSec_CAF1 67944 108 + 1 UUUAAUUGACAUUUUGUGUCGUGGCUUUUGUUUUGAAAUACCGAGUAUAUGGCCUUAAUUGGGCUCAUAUG-UGGUAAAUAAGUCAUUUCAGAUUUCUUGGAAAAG-----------AGA .......(((((...)))))(((((((...........(((((..((((((((((.....)))).))))))-)))))...)))))))......((((((....)))-----------))) ( -26.34) >DroSim_CAF1 69075 109 + 1 UUUAAUUGACAUUUUGUGUCGUGGCUUUUGUUUUGAAAUACCGAGUAUAUGGACUUAAUUGGGCUCAUAUGCUGGUAAAUAAGUCGUUUCAGAUUUCUUGGAAAAG-----------AGA .......(((........(((..((....))..)))..((((.((((((((..(((....)))..)))))))))))).....)))........((((((....)))-----------))) ( -26.40) >DroEre_CAF1 85978 120 + 1 UUUAAUUGACAUUUUGUGUCGGGGCUUUUGUUUUGAAAUACCCAAUAUAUGGCCUUAAUUGGACUCAUAUGUUGGUAAAUAAAUCGUUUCAGAUUUCGUGGAAAAGUAGACUUACACAAA ............(((((((.((.((((((.(..(((((...(((((((((((((......)).).))))))))))........((......))))))).).))))))...)).))))))) ( -29.50) >DroYak_CAF1 70949 120 + 1 UUUAAUUGACAUUUUGUGUCGGGGCUUUUGUUUUGAAAUACCGAAUAUAUAGCCUUAAUUGGGCUCAUAUGUUGGUAAAUAAGUCGUUUCAGAUUUCGUAGAAAUGUAGAUGUACGGAAA .......(((........(((((((....)))))))..((((.((((((((((((.....))))).))))))))))).....)))........(((((((.(........).))))))). ( -29.70) >consensus UUUAAUUGACAUUUUGUGUCGUGGCUUUUGUUUUGAAAUACCGAGUAUAUGGCCUUAAUUGGGCUCAUAUGUUGGUAAAUAAGUCGUUUCAGAUUUCUUGGAAAAG___________AGA ......((((((...)))))).((..((((...(((..((((.((((((((((((.....)))).))))))))))))......)))...))))..))....................... (-18.54 = -18.78 + 0.24)


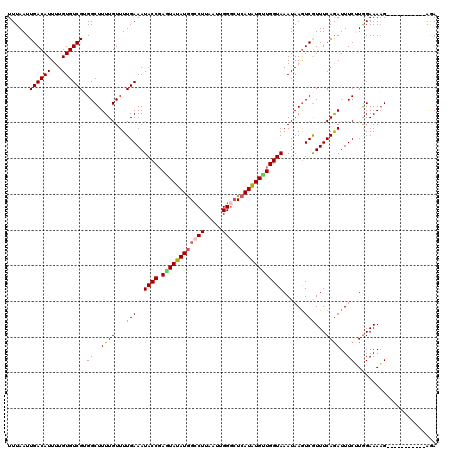
| Location | 3,537,819 – 3,537,928 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -13.14 |
| Energy contribution | -14.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3537819 109 + 23771897 CCGAGUAUAUGGCCUUAAUUGGGAUCAUGUGUUGGUAAAUAAGUCGUUUCAAAUUUCUUGGAAAAG-----------AGAAAAAUUUGGUGACCAGGAAAUAGUUAAAUUCAGACUACAA ((((.((((((((((.....))).)))))))))))......(((((((((...((((((....)))-----------))).....((((...))))))))).(.......).)))).... ( -22.80) >DroSec_CAF1 67984 107 + 1 CCGAGUAUAUGGCCUUAAUUGGGCUCAUAUG-UGGUAAAUAAGUCAUUUCAGAUUUCUUGGAAAAG-----------AGAA-AAUUGGGUGGCCAGGAAAUACUUAAAUUUAAACUACCA (((..((((((((((.....)))).))))))-)))....(((((.(((((...((((((....)))-----------))).-..((((....)))))))))))))).............. ( -26.20) >DroSim_CAF1 69115 109 + 1 CCGAGUAUAUGGACUUAAUUGGGCUCAUAUGCUGGUAAAUAAGUCGUUUCAGAUUUCUUGGAAAAG-----------AGAAAAAUUGGGCGACCAGGAAAUAGUUAAAUUCAAACUACCA ...((((((((..(((....)))..))))))))((((........(((((...((((((....)))-----------)))....((((....))))))))).(((.......))))))). ( -27.50) >DroEre_CAF1 86018 120 + 1 CCCAAUAUAUGGCCUUAAUUGGACUCAUAUGUUGGUAAAUAAAUCGUUUCAGAUUUCGUGGAAAAGUAGACUUACACAAAAUAAUUGCUGAACCAGGAAAUAGUGAAAUUUAAACAACCA .(((((((((((((......)).).))))))))))..........((((((.(((((.(((...(((((...............)))))...))).)))))..))))))........... ( -28.66) >DroYak_CAF1 70989 104 + 1 CCGAAUAUAUAGCCUUAAUUGGGCUCAUAUGUUGGUAAAUAAGUCGUUUCAGAUUUCGUAGAAAUGUAGAUGUACGGAAAAUAAUUAAAUGACCAGGAAGUAGU---------------- ((.((((((((((((.....))))).))))))).........(((((((....(((((((.(........).))))))).......)))))))..)).......---------------- ( -22.10) >consensus CCGAGUAUAUGGCCUUAAUUGGGCUCAUAUGUUGGUAAAUAAGUCGUUUCAGAUUUCUUGGAAAAG___________AGAAAAAUUGGGUGACCAGGAAAUAGUUAAAUUCAAACUACCA ((.((((((((((((.....)))).)))))))))).................(((((((((...............................)))))))))................... (-13.14 = -14.14 + 1.00)

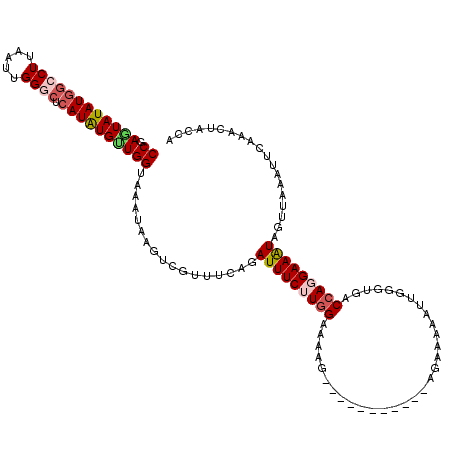
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:58 2006