
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,532,085 – 3,532,213 |
| Length | 128 |
| Max. P | 0.938049 |

| Location | 3,532,085 – 3,532,191 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -27.68 |
| Energy contribution | -28.16 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3532085 106 + 23771897 UUUGCCUGGUGAAUAAAUGAUGCGUGUUUUAUGGCGACGCUGAUGGAUGUGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCAGCCAGA-ACUAG .....(((((...........(((((((....))).)))).......(.(((((((((.((((.......))))((((......))))..))))))))).)-))))) ( -33.70) >DroSec_CAF1 62297 106 + 1 UUUGUGUGGUGAAUAAAUGAUGCGUGUUUUAUGGCGACGCUGAUGGAUGUGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGCUUAAUGGCAGCCAGA-ACUAA ...((((.((..(((((..........))))).)).))))...(((.(.(((((((((.((((.......))))((((.....))))...))))))))).)-.))). ( -35.50) >DroSim_CAF1 63334 106 + 1 UUUGCCUGGUGAAUAAAUGAUGCGUGUUUUAUGGCGACGCUGAUGGAUGUGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCAGCCAGA-ACUAA .(((((..(((((...(((...)))..))))))))))......(((.(.(((((((((.((((.......))))((((......))))..))))))))).)-.))). ( -32.50) >DroEre_CAF1 80118 107 + 1 UUUGCCUGGUGAAUAAAUGAUGCGUGUUUUAUAGCGACGCUGAUGGAUGGGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCACCCAGAUAUUAG .....................(((((((....))).))))(((((..((((..(((((.((((.......))))((((......))))..)))))))))..))))). ( -28.20) >DroYak_CAF1 65052 106 + 1 UUUGCCUGGUGAAUAAAUGAUGCGUGUUUUAUGGCGACGCUGAUGGAUGGGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCAGCCAGA-ACUAG .....(((((...........(((((((....))).))))..........((((((((.((((.......))))((((......))))..))))))))...-))))) ( -32.30) >consensus UUUGCCUGGUGAAUAAAUGAUGCGUGUUUUAUGGCGACGCUGAUGGAUGUGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCAGCCAGA_ACUAG .....(((((...........(((((((....))).))))..........((((((((.((((.......))))((((......))))..))))))))....))))) (-27.68 = -28.16 + 0.48)

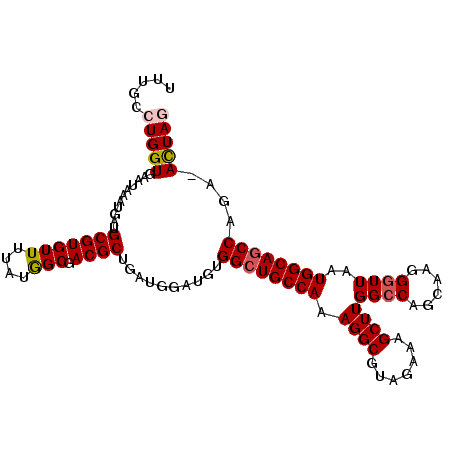
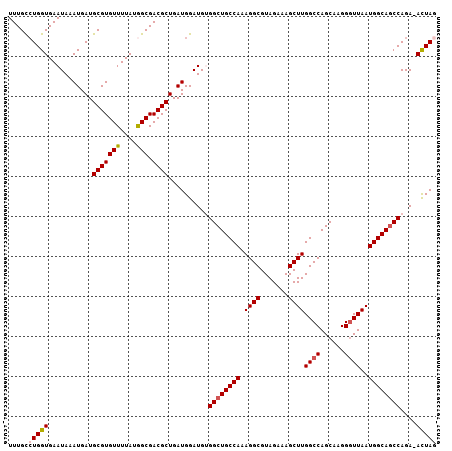
| Location | 3,532,085 – 3,532,191 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -20.64 |
| Energy contribution | -20.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3532085 106 - 23771897 CUAGU-UCUGGCUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCACAUCCAUCAGCGUCGCCAUAAAACACGCAUCAUUUAUUCACCAGGCAAA ...((-(.(((((((((.........(((.....)))............))))))))).........((((.(........)))))...............)))... ( -23.00) >DroSec_CAF1 62297 106 - 1 UUAGU-UCUGGCUGCCAUUAAGCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCACAUCCAUCAGCGUCGCCAUAAAACACGCAUCAUUUAUUCACCACACAAA .....-..(((((((((....(((.....)))...((.......))...))))))))).........((((.(........)))))..................... ( -25.30) >DroSim_CAF1 63334 106 - 1 UUAGU-UCUGGCUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCACAUCCAUCAGCGUCGCCAUAAAACACGCAUCAUUUAUUCACCAGGCAAA ...((-(.(((((((((.........(((.....)))............))))))))).........((((.(........)))))...............)))... ( -23.00) >DroEre_CAF1 80118 107 - 1 CUAAUAUCUGGGUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCCCAUCCAUCAGCGUCGCUAUAAAACACGCAUCAUUUAUUCACCAGGCAAA .........((((.......))))(((((.(((((((.......))..)))))..............((((.(........)))))...............))))). ( -21.00) >DroYak_CAF1 65052 106 - 1 CUAGU-UCUGGCUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCCCAUCCAUCAGCGUCGCCAUAAAACACGCAUCAUUUAUUCACCAGGCAAA ...((-(..((((((((.........(((.....)))............))))))))..........((((.(........)))))...............)))... ( -21.60) >consensus CUAGU_UCUGGCUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCACAUCCAUCAGCGUCGCCAUAAAACACGCAUCAUUUAUUCACCAGGCAAA .........((((((((.........(((.....)))............))))))))..........((((.(........)))))..................... (-20.64 = -20.84 + 0.20)


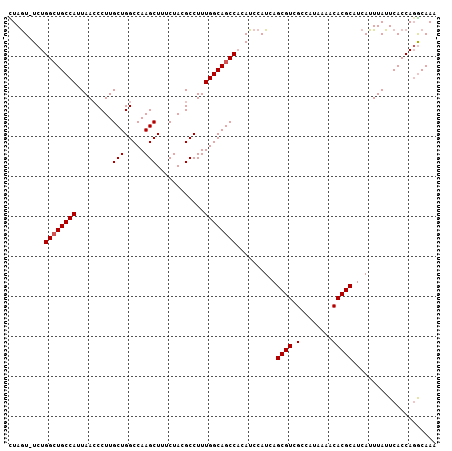
| Location | 3,532,115 – 3,532,213 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -23.36 |
| Energy contribution | -23.44 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3532115 98 + 23771897 AUGGCGACGCUGAUGGAUGUGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCAGCCAGA-ACUAGA---------AAUUUAUGUUUGCGUGAAGUU ......((((...(((.(.(((((((((.((((.......))))((((......))))..))))))))).)-.))).(---------(((....))))))))...... ( -30.60) >DroSec_CAF1 62327 98 + 1 AUGGCGACGCUGAUGGAUGUGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGCUUAAUGGCAGCCAGA-ACUAAA---------AAUUUAUGUUUGCGUGAAGUU ......((((...(((.(.(((((((((.((((.......))))((((.....))))...))))))))).)-.))).(---------(((....))))))))...... ( -32.30) >DroSim_CAF1 63364 98 + 1 AUGGCGACGCUGAUGGAUGUGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCAGCCAGA-ACUAAA---------AAUUUAUGUUUGCGUGAAGUU ......((((...(((.(.(((((((((.((((.......))))((((......))))..))))))))).)-.))).(---------(((....))))))))...... ( -30.50) >DroEre_CAF1 80148 108 + 1 AUAGCGACGCUGAUGGAUGGGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCACCCAGAUAUUAGAAAUAGUCGGAAUUUAUGUUUGCGUGAAAUU ....((((.((((((..((((..(((((.((((.......))))((((......))))..)))))))))..)))))).....))))...((((((....))))))... ( -34.00) >DroYak_CAF1 65082 98 + 1 AUGGCGACGCUGAUGGAUGGGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCAGCCAGA-ACUAGA---------CAUUUGUGUUUACGUGAAGUU (((..(((((.((((..(((((((((((.((((.......))))((((......))))..))))))))...-.)))..---------)))).)))))..)))...... ( -34.60) >consensus AUGGCGACGCUGAUGGAUGUGGCUGCCAAAGGCGUAGAAAGCUUGGCCAGCAAGGGUUAAUGGCAGCCAGA_ACUAGA_________AAUUUAUGUUUGCGUGAAGUU .((((...))))........((((((((.((((.......))))((((......))))..)))))))).....................((((((....))))))... (-23.36 = -23.44 + 0.08)

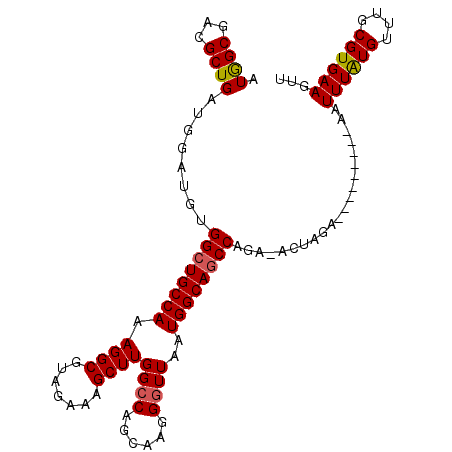

| Location | 3,532,115 – 3,532,213 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -17.92 |
| Energy contribution | -18.16 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3532115 98 - 23771897 AACUUCACGCAAACAUAAAUU---------UCUAGU-UCUGGCUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCACAUCCAUCAGCGUCGCCAU ......((((.(((.......---------....))-).(((((((((.........(((.....)))............))))))))).........))))...... ( -22.20) >DroSec_CAF1 62327 98 - 1 AACUUCACGCAAACAUAAAUU---------UUUAGU-UCUGGCUGCCAUUAAGCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCACAUCCAUCAGCGUCGCCAU ......((((.(((.(((...---------.)))))-).(((((((((....(((.....)))...((.......))...))))))))).........))))...... ( -25.00) >DroSim_CAF1 63364 98 - 1 AACUUCACGCAAACAUAAAUU---------UUUAGU-UCUGGCUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCACAUCCAUCAGCGUCGCCAU ......((((.(((.(((...---------.)))))-).(((((((((.........(((.....)))............))))))))).........))))...... ( -22.50) >DroEre_CAF1 80148 108 - 1 AAUUUCACGCAAACAUAAAUUCCGACUAUUUCUAAUAUCUGGGUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCCCAUCCAUCAGCGUCGCUAU ......((((..............................((((.......))))..((((.((((((.......))..))))))))...........))))...... ( -20.60) >DroYak_CAF1 65082 98 - 1 AACUUCACGUAAACACAAAUG---------UCUAGU-UCUGGCUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCCCAUCCAUCAGCGUCGCCAU ((((..((((........)))---------)..)))-)..((((((((.........(((.....)))............)))))))).................... ( -20.50) >consensus AACUUCACGCAAACAUAAAUU_________UCUAGU_UCUGGCUGCCAUUAACCCUUGCUGGCCAAGCUUUCUACGCCUUUGGCAGCCACAUCCAUCAGCGUCGCCAU ......((((.......................((...))((((((((.........(((.....)))............))))))))..........))))...... (-17.92 = -18.16 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:53 2006