
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,529,611 – 3,529,721 |
| Length | 110 |
| Max. P | 0.602924 |

| Location | 3,529,611 – 3,529,721 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -16.05 |
| Energy contribution | -16.42 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3529611 110 + 23771897 GCCGAAUUAAUCAGUCAGCCCACUAAAAUUAACAACAAUAAUUUUGGUGCCGGUGCGUAUAUACACAUCAUAAAUAAUCCACAUAACAUGACUGUAUCUUUUUUGUUUGA ..((((.....((((((...(((((((((((.......)))))))))))..((((.((....)).))))...................)))))).......))))..... ( -21.40) >DroSec_CAF1 59917 109 + 1 GCCGAAUUGAUCAGUCAGCCCACUAAAAUUAACAACAAUAAUUUUGGUGCCGGUGCGUAUAUACACAUCAUAAAUAAUCCACAUAACAUGACUGUAUCUU-UUUGUUUGA ..((((..(((((((((...(((((((((((.......)))))))))))..((((.((....)).))))...................)))))).)))..-))))..... ( -26.40) >DroEre_CAF1 77669 109 + 1 GCCGAAUUAAUCAGUCAGCCCACUAAAAUUAACAACAAUAAUUUUGGUGCCGGUGCGUAUAUACACAUCAUAAAUAAUCCACAUAACAUGACUGUAUCUU-UUUGUUUGA ..((((.....((((((...(((((((((((.......)))))))))))..((((.((....)).))))...................))))))......-))))..... ( -22.00) >DroWil_CAF1 101689 91 + 1 GCCGAAUUAAUAAAUCAACCAACUAAAAUUAACAACAACAAUUUUGGUGCCGGUGCGUAUAAACA--UUA-AAAUUAUGUACAUAGUUUAAUUU---------------- ...((((((............(((((((((.........)))))))))....(((((((......--...-....))))))).)))))).....---------------- ( -12.22) >DroYak_CAF1 62610 109 + 1 GCCGAAUUAAUCAGUCAGCCCACUAAAAUUAACAACAAUAAUUUUGGUGCCGGUGCGUAUAUACACAUCAUAAAUAAUCCACAUAACAUGACUGUAUCUU-UUUGUUUGA ..((((.....((((((...(((((((((((.......)))))))))))..((((.((....)).))))...................))))))......-))))..... ( -22.00) >DroAna_CAF1 73869 107 + 1 GCCGAACUAAUCAGUCAGCCCACUAAAAUUAACAACAAUAAUUUUGGUGCCGGUGCGUAUAUACACAUCAUAAAUAAUCCCCCCAUC-UGACUACA-CUU-AUUGUUUGA ..(((((.....((((((..(((((((((((.......)))))))))))..((((.((....)).)))).................)-)))))...-...-...))))). ( -22.14) >consensus GCCGAAUUAAUCAGUCAGCCCACUAAAAUUAACAACAAUAAUUUUGGUGCCGGUGCGUAUAUACACAUCAUAAAUAAUCCACAUAACAUGACUGUAUCUU_UUUGUUUGA ...........((((((...(((((((((((.......)))))))))))..((((.((....)).))))...................))))))................ (-16.05 = -16.42 + 0.36)

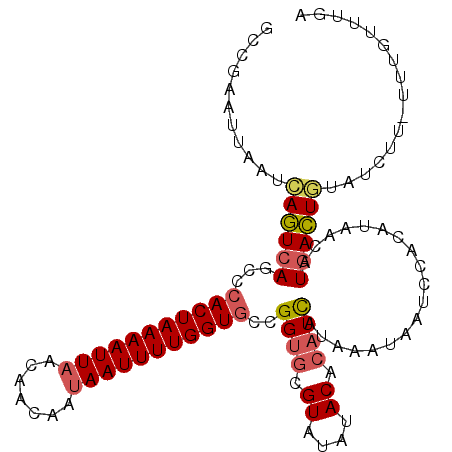
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:49 2006