| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
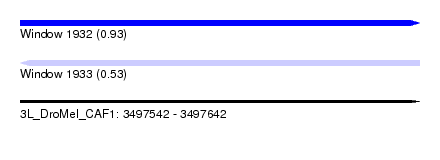
| Location | 3,497,542 – 3,497,642 |
| Length | 100 |
| Max. P | 0.925483 |

| Location | 3,497,542 – 3,497,642 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
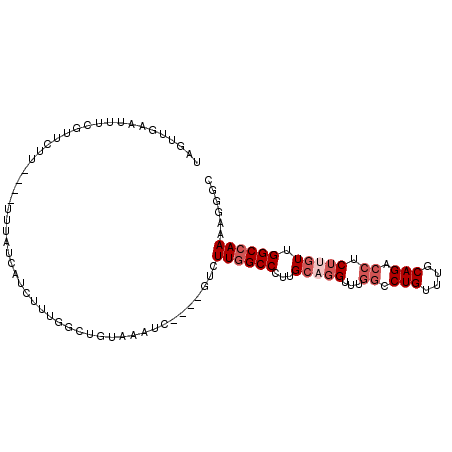
>3L_DroMel_CAF1 3497542 100 + 23771897 GCCCUUUUGGCCAACAAGAGGUCUGCAAACAGGCCAAACCUGCAAGGGCCAAGAC----GAUUUACAACCAAAGAUGAUAAAAAAAAAGAACGAAAUUCAACUA ..(.((((((((....((.((((((....))))))....)).....)))))))).----)............................................ ( -19.60) >DroSec_CAF1 28011 98 + 1 GCCCUUUUGGCCAACAAGAGGUCUGCAAACAGGCCAAACCAGCAAGGGCCAAGAC----GAUUUACAACCAAAGAUGAUAAAA--AAAGAAUGAAAUUCAACUU ((((((((((.(.....).((((((....))))))...)))).))))))......----............(((.(((.....--............))).))) ( -20.03) >DroSim_CAF1 28396 96 + 1 GCCCUUUUGGCCAACA-GAGGUCUGCAAACAGGCCAAACCUGCAAGGGCCAAGAC----GAUUUACAACCAAAGAUGAUAAA---AAAGAACGAAAUUCAACUA ..(.((((((((..((-(.((((((....))))))....)))....)))))))).----)............((.(((....---............))).)). ( -21.29) >DroEre_CAF1 43782 96 + 1 GCUCUUUUGGCCAACAAGAGGCCUGCAAACAGGCCAAACCUGCAAGGGCCAAGUC----GAUUUACAGCCAAAGGUAAUAAA----AAGAAUACAAUUCAAGUA (((..(((((((....((.((((((....))))))....)).....)))))))((----..((((..(((...)))..))))----..))..........))). ( -23.70) >DroYak_CAF1 29478 96 + 1 GCCCUUUUGGCCAACAAGAGGUCUGCAAACAGGCCAAACCUGCAAGGGCCAAGAC----GAUUUACAGCCAAAGAUGAUAAA----AAGAACACAAAUCGAGGA ..((((((((((....((.((((((....))))))....)).....))))))))(----(((((..................----........)))))).)). ( -22.97) >DroAna_CAF1 42395 82 + 1 GCCCUUUUGGCCAAGAAGAUCUCUGCAAACAGGCCCCACCAGCAG-GGCUAAAAAGCAGGGCUUAAAG--AAAGAAAUUAAA----AAG--------------- ...((((.((((.((.....))((((.....(((((........)-)))).....)))))))).))))--............----...--------------- ( -21.30) >consensus GCCCUUUUGGCCAACAAGAGGUCUGCAAACAGGCCAAACCUGCAAGGGCCAAGAC____GAUUUACAACCAAAGAUGAUAAA____AAGAACGAAAUUCAACUA ....((((((((.......((((((....))))))...........)))))))).................................................. (-16.37 = -16.45 + 0.08)



| Location | 3,497,542 – 3,497,642 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.82 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3497542 100 - 23771897 UAGUUGAAUUUCGUUCUUUUUUUUUAUCAUCUUUGGUUGUAAAUC----GUCUUGGCCCUUGCAGGUUUGGCCUGUUUGCAGACCUCUUGUUGGCCAAAAGGGC ............((((((((..(((((.((.....)).)))))..----.....((((...(((((...((.(((....))).)).))))).)))))))))))) ( -24.20) >DroSec_CAF1 28011 98 - 1 AAGUUGAAUUUCAUUCUUU--UUUUAUCAUCUUUGGUUGUAAAUC----GUCUUGGCCCUUGCUGGUUUGGCCUGUUUGCAGACCUCUUGUUGGCCAAAAGGGC (((.(((.((.((..(...--.............)..)).)).))----).))).((((((..(((((.((.(((....))).)).......))))).)))))) ( -19.39) >DroSim_CAF1 28396 96 - 1 UAGUUGAAUUUCGUUCUUU---UUUAUCAUCUUUGGUUGUAAAUC----GUCUUGGCCCUUGCAGGUUUGGCCUGUUUGCAGACCUC-UGUUGGCCAAAAGGGC ............(((((((---(((((.((.....)).)))))..----....(((((...(((((.....)))))..((((....)-))).)))))))))))) ( -22.80) >DroEre_CAF1 43782 96 - 1 UACUUGAAUUGUAUUCUU----UUUAUUACCUUUGGCUGUAAAUC----GACUUGGCCCUUGCAGGUUUGGCCUGUUUGCAGGCCUCUUGUUGGCCAAAAGAGC ..(((.........((..----(((((..((...))..)))))..----)).((((((...(((((...((((((....)))))).))))).)))))))))... ( -30.70) >DroYak_CAF1 29478 96 - 1 UCCUCGAUUUGUGUUCUU----UUUAUCAUCUUUGGCUGUAAAUC----GUCUUGGCCCUUGCAGGUUUGGCCUGUUUGCAGACCUCUUGUUGGCCAAAAGGGC ((((((((((..(..(..----............)..)..)))))----)..((((((...(((((...((.(((....))).)).))))).)))))).)))). ( -26.34) >DroAna_CAF1 42395 82 - 1 ---------------CUU----UUUAAUUUCUUU--CUUUAAGCCCUGCUUUUUAGCC-CUGCUGGUGGGGCCUGUUUGCAGAGAUCUUCUUGGCCAAAAGGGC ---------------...----..........((--((((..(((((((......(((-(((....))))))......))))((.....)).)))..)))))). ( -20.40) >consensus UAGUUGAAUUUCGUUCUU____UUUAUCAUCUUUGGCUGUAAAUC____GUCUUGGCCCUUGCAGGUUUGGCCUGUUUGCAGACCUCUUGUUGGCCAAAAGGGC ....................................................((((((...(((((...((.(((....))).)).))))).))))))...... (-15.65 = -16.82 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:41 2006