| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 405,343 – 405,434 |
| Length | 91 |
| Max. P | 0.579001 |

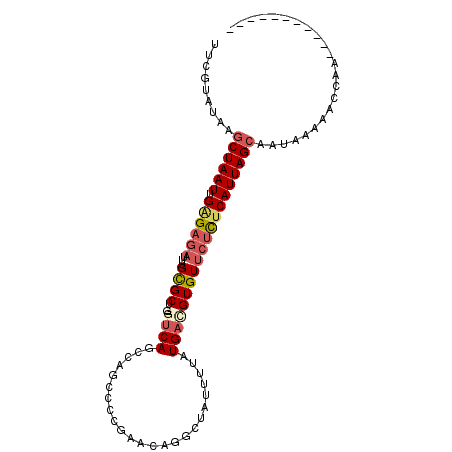
| Location | 405,343 – 405,434 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -13.24 |
| Energy contribution | -14.82 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
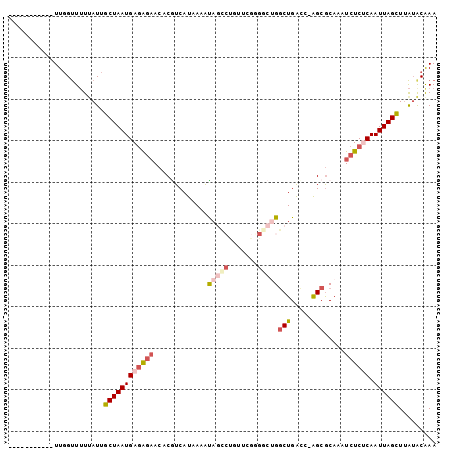
>3L_DroMel_CAF1 405343 91 + 23771897 -----------UUGGUUUUUAUUGCUAAUGAGAGAACACGUCAUAAAAUAGCCUGUUCGGGGCUGGCUGACC-AGCGCAAAUCUCUCAAUUAGCUUAUACGAA -----------............((((((((((((................(((....)))(((((....))-))).....)))))).))))))......... ( -26.30) >DroGri_CAF1 62170 97 + 1 UGUGUGUGUUAUUCGUUUUAAUUGCUAAUGAGAG-ACACAUCAUAAAAUAACACAU-CAU----GCCUGGCCAGGCGCAAAUCUCUCAAUUAGUUUAUACAAA ((((((((((.((((((.........)))))).)-))))).))))...........-...----((((....))))........................... ( -18.60) >DroSec_CAF1 52306 91 + 1 -----------UUGGUUUUUAUUGCUAAUGAGAGAACACGUCAUAAAAUAGCCCAGUCGGGGCUGGCUGACC-AGCGCAAAUCUCUCAAUUAGCUUAUACGAA -----------............((((((((((((.............((((((.....))))))(((....-))).....)))))).))))))......... ( -28.00) >DroEre_CAF1 56703 90 + 1 -----------UUGGUUUUUAUUGCUAAUGAGAGAACACGUCAUAAAGCAGUCUGUGCG-GGCUGGCUGAUC-AGCGCAAAUCUCCCAAUUAGCUUAUGCGAA -----------............((((((..((((...(((.....((((((((....)-)))).)))....-.)))....))))...))))))......... ( -21.30) >DroYak_CAF1 56636 91 + 1 -----------UUGGUUUUUAUUGCUAAUGAGAGAACACGUCAUAAAAUAGCCUGUUCGGGGCUGGCUGAUC-AGCGCAAAUCUCGCAAUUAGCUUAUACAAA -----------............((((((..((((.............((((((.....))))))(((....-))).....))))...))))))......... ( -22.10) >DroAna_CAF1 46471 80 + 1 -----------UUGGCU-UUAUUGCUAAUGAAGGAACACAUCAUAAAGUUUC------CG----GGCUGACC-AGCACAAAUAUCUCAAUUAGCUCGAACAAA -----------(((((.-.....)))))....((((.((........)))))------)(----((((((..-................)))))))....... ( -14.17) >consensus ___________UUGGUUUUUAUUGCUAAUGAGAGAACACGUCAUAAAAUAGCCUGUUCGGGGCUGGCUGACC_AGCGCAAAUCUCUCAAUUAGCUUAUACAAA .......................((((((((((((.............(((((.......)))))(((.....))).....)))))).))))))......... (-13.24 = -14.82 + 1.57)

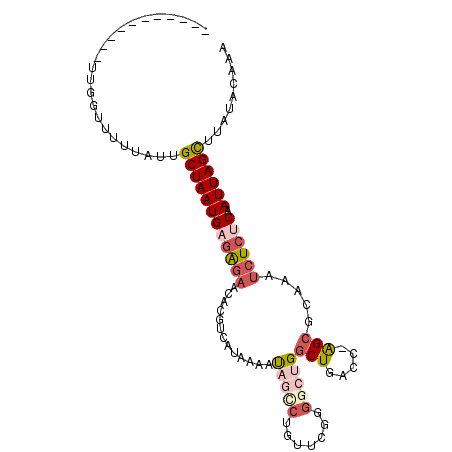
| Location | 405,343 – 405,434 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -12.37 |
| Energy contribution | -13.21 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 405343 91 - 23771897 UUCGUAUAAGCUAAUUGAGAGAUUUGCGCU-GGUCAGCCAGCCCCGAACAGGCUAUUUUAUGACGUGUUCUCUCAUUAGCAAUAAAAACCAA----------- .........((((((.((((((..((((((-((....))))).((.....))............))).))))))))))))............----------- ( -26.50) >DroGri_CAF1 62170 97 - 1 UUUGUAUAAACUAAUUGAGAGAUUUGCGCCUGGCCAGGC----AUG-AUGUGUUAUUUUAUGAUGUGU-CUCUCAUUAGCAAUUAAAACGAAUAACACACACA (((((.....(((((.(((((((..((((((....))))----((.-(((........))).))))))-))))))))))........)))))........... ( -19.22) >DroSec_CAF1 52306 91 - 1 UUCGUAUAAGCUAAUUGAGAGAUUUGCGCU-GGUCAGCCAGCCCCGACUGGGCUAUUUUAUGACGUGUUCUCUCAUUAGCAAUAAAAACCAA----------- .........((((((.((((((...((((.-.((((...(((((.....)))))......))))))))))))))))))))............----------- ( -29.60) >DroEre_CAF1 56703 90 - 1 UUCGCAUAAGCUAAUUGGGAGAUUUGCGCU-GAUCAGCCAGCC-CGCACAGACUGCUUUAUGACGUGUUCUCUCAUUAGCAAUAAAAACCAA----------- .........((((((.((((((..((((((-(......)))).-.(((.....)))........))).))))))))))))............----------- ( -22.80) >DroYak_CAF1 56636 91 - 1 UUUGUAUAAGCUAAUUGCGAGAUUUGCGCU-GAUCAGCCAGCCCCGAACAGGCUAUUUUAUGACGUGUUCUCUCAUUAGCAAUAAAAACCAA----------- .........((((((...((((.....(((-(......))))...(((((.((........).).)))))))))))))))............----------- ( -20.40) >DroAna_CAF1 46471 80 - 1 UUUGUUCGAGCUAAUUGAGAUAUUUGUGCU-GGUCAGCC----CG------GAAACUUUAUGAUGUGUUCCUUCAUUAGCAAUAA-AGCCAA----------- .........((((((.(((......(.(((-....))).----)(------(((((........)).))))))))))))).....-......----------- ( -13.90) >consensus UUCGUAUAAGCUAAUUGAGAGAUUUGCGCU_GGUCAGCCAGCCCCGAACAGGCUAUUUUAUGACGUGUUCUCUCAUUAGCAAUAAAAACCAA___________ .........((((((.((((((...((((...((((........................))))))))))))))))))))....................... (-12.37 = -13.21 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:04 2006