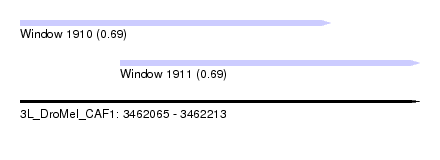
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,462,065 – 3,462,213 |
| Length | 148 |
| Max. P | 0.689982 |

| Location | 3,462,065 – 3,462,180 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -19.80 |
| Energy contribution | -21.37 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
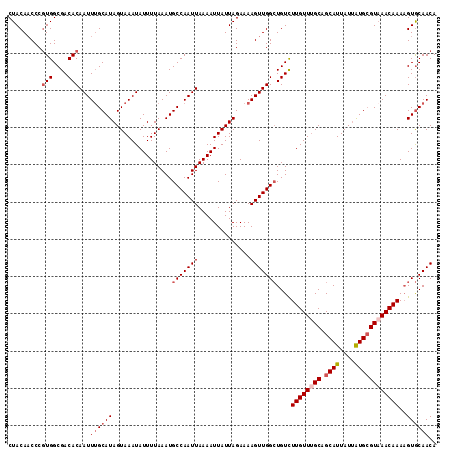
>3L_DroMel_CAF1 3462065 115 + 23771897 CUACAACCCGUGGCGACACAAUUUGCAUAGUAAAUAUUUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGCUGUCUUGUUUGCAGCAUUAUUAUGAGUAAACAAAAGUGCAACA .........(((....)))...((((((................(((((((..............)))))))....((((((((..(((....))).))))))))..)))))).. ( -24.34) >DroSec_CAF1 9379 115 + 1 CUACAACCCGUGGCGACACAAUUAGCAUAGUAAAUAUUUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGCUGUCUUGUUUGCAGCAUUAUUAUGCGUAAACAAAAGUGCAACA .........(((....))).....((((................(((((((..............)))))))....((((((((.((((....))))))))))))..)))).... ( -27.14) >DroSim_CAF1 9402 112 + 1 CUACA-CCCGUGGCGACACAAUUUGCAUAGUAAAUAUUUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGCUGUCUUGUU-GC-GCAUUAUUAUGCGUAAACAAAAGUGCAACA .....-...(((....)))...((((((................(((((((..............)))))))....(((((-((-((((....)))))).)))))..)))))).. ( -27.54) >DroYak_CAF1 9646 115 + 1 CUACAACCCAUGGCGACACAAUUUGCAUAGUAAAUAUUUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGAUGUUUUGUUUGCAGCAUUAUCGUGCGUCAACAAAUGUACAACA .((((.....(((((.(((.((.(((...(((((((....((...((((((..............))))))...)).))))))).)))..)).)))))))).....))))..... ( -21.04) >consensus CUACAACCCGUGGCGACACAAUUUGCAUAGUAAAUAUUUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGCUGUCUUGUUUGCAGCAUUAUUAUGCGUAAACAAAAGUGCAACA .........(((....)))...((((((................(((((((..............)))))))....((((((((.((((....))))))))))))..)))))).. (-19.80 = -21.37 + 1.56)

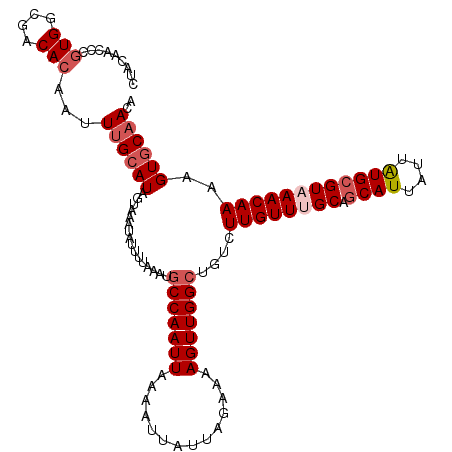
| Location | 3,462,102 – 3,462,213 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.39 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.54 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
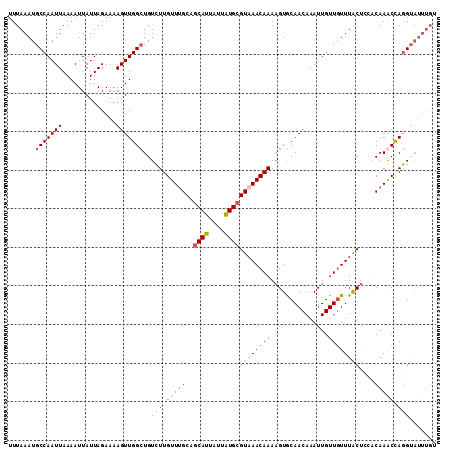
>3L_DroMel_CAF1 3462102 111 + 23771897 UUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGCUGUCUUGUUUGCAGCAUUAUUAUGAGUAAACAAAAGUGCAACAAAUUGUUGUUUACUCCACAAACUAGGUAUUUGU ..((((((((.......................(((((.......)))))........((((((((((.(((.......))).)))))))))).........)))))))). ( -25.80) >DroSec_CAF1 9416 111 + 1 UUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGCUGUCUUGUUUGCAGCAUUAUUAUGCGUAAACAAAAGUGCAACAAAUUGUUGUUUACUCUCCAAGCCAGGUAUUUGU ..((((((((..((((.....))))......(((((...((((((((.((((....)))))))))))).((((((((.....)))))..))).....))))))))))))). ( -29.30) >DroSim_CAF1 9438 108 + 1 UUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGCUGUCUUGUU-GC-GCAUUAUUAUGCGUAAACAAAAGUGCAACA-AUUGUUGUUUACUUUCCAAGCCAGGUAUUUGU ..((((((((..((((.....))))......(((((....((((-((-((((....)))))).))))((((((((((.-...)))))..)))))...))))))))))))). ( -29.40) >DroYak_CAF1 9683 104 + 1 UUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGAUGUUUUGUUUGCAGCAUUAUCGUGCGUCAACAAAUGUACAACAAAUUGUUAUUUGCUACACAAACUAAG------- ...(((((((((((..............)))))).))))).(((((.((((.....((((((......))))))(((.....)))...))))...)))))....------- ( -15.64) >consensus UUUAAAUGCCAAUUAAAAUUAUUAGAAAAGUUGGCUGUCUUGUUUGCAGCAUUAUUAUGCGUAAACAAAAGUGCAACAAAUUGUUGUUUACUCCACAAACCAGGUAUUUGU .......(((((((..............)))))))..(((.(((((..((((....))))((((((((.(((.......))).))))))))....))))).)))....... (-16.04 = -16.54 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:20 2006